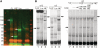Identification of CTCF as a master regulator of the clustered protocadherin genes - PubMed (original) (raw)
Identification of CTCF as a master regulator of the clustered protocadherin genes
Michal Golan-Mashiach et al. Nucleic Acids Res. 2012 Apr.
Abstract
The brain is a large and complex network of neurons. Specific neuronal connectivity is thought to be based on the combinatorial expression of the 52 protocadherins (Pcdh) membrane adhesion proteins, whereby each neuron expresses only a specific subset. Pcdh genes are arranged in tandem, in a cluster of three families: Pcdhα, Pcdhβ and Pcdhγ. The expression of each Pcdh gene is regulated by a promoter that has a regulatory conserved sequence element (CSE), common to all 52 genes. The mechanism and factors controlling individual Pcdh gene expression are currently unknown. Here we show that the promoter of each Pcdh gene contains a gene-specific conserved control region, termed specific sequence element (SSE), located adjacent and upstream to the CSE and activates transcription together with the CSE. We purified the complex that specifically binds the SSE-CSE region and identified the CCTC binding-factor (CTCF) as a key molecule that binds and activates Pcdh promoters. Our findings point to CTCF as a factor essential for Pcdh expression and probably governing neuronal connectivity.
Figures
Figure 1.
Schematic representation of the Pcdh gene cluster promoters. Genomic organization of the Pcdh genes, with tandem variable region exons (blue), promoter regions (turquoise) and constant region exons (red). Each promoter region contains an SSE followed by a CSE, which is common to all genes of the family. V, variable region; C, constant region.
Figure 2.
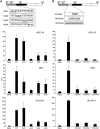
αSSE and CSE activate transcription cooperatively. (A) The expression profile of all the Pcdhα genes in HEC-1B, 293T and SH-SY5Y cell lines by RT–PCR, (
Supplementary Figure S2
). Gray and black boxes indicate for absence or presence of Pcdhα mRNA, respectively. (B) A scheme depicting the sequences of α6SSE–CSE WT and mutants (b–d). (C) WT (α6SSE–CSE and α3SSE–CSE) and the mutated promoters (fused to firefly luciferase reporter gene) were transfected into HEC-1B, 293T and SH-SY5Ycell lines together with RSV-Renilla that serves as internal control. The parental pGL3-basic (Basic) was also transfected as a control. Twenty-four hours post-transfection firefly and renilla luciferase activities were measured. The normalized results are the mean of at least four independent experiments (±SD).
Figure 3.
Sequence requirements for the function of α6SSE–CSE as transcriptional elements. (A) Successive blocks within αSSE (underlined) in the α6SSE–CSE promoter were mutated (Mut1-Mut4). The wild-type and mutated constructs were transfected into HEC-1B, 293T and SH-SY5Y cell lines together with RSV-Renilla that serves as internal control. Twenty-four hours post-transfection firefly and Renilla luciferase activities were measured. The normalized results are the mean of at least four independent experiments (±SD). (B) Linkers of 5, 10 and 15 bp (5nLinker, 10nLinker and 5nLinker) were introduced between α6SSE and CSE and their effect was analyzed as described earlier.
Figure 4.
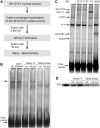
Gene-specific complex binds to the αSSE–CSE. (A) EMSA using HEC-1B cell nuclear extract and a fluorescently labeled double stranded oligonucleotide containing αSSE–CSE sequence as a probe. Lane 1, the probe is incubated with HEC-1B nuclear extract. Competitor DNAs were added to the reactions in lanes 2 and 3 as indicated on the top. The sequences of the oligonucleotides used for binding and competition are shown on the top. (B) αSSE–CSE display cooperative DNA binding activity. EMSA using HEC-1B cell nuclear extract and a fluorescently labeled double stranded oligonucleotide containing α6SSE–CSE as a probe. Unlabeled competitor DNAs were added to the reactions as indicated in the top panel. The sequences of the oligonucleotides used for binding and competition are shown earlier. (C) The specific complex is shown to bind with high affinity to α6SSE–CSE. EMSA using SH-SY5Y cell nuclear extract and a fluorescently labeled double stranded oligonucleotide containing α6SSE–CSE as a probe. Unlabeled αSSE–CSE sequences of Pcdhα genes as competitor DNAs were added in excess to the reactions as indicated in the top panel. The sequences of the oligonucleotides used for binding and competition are shown earlier. (D) Competition assay for α3SSE–CSE and to α12SSE–CSE specific complex. EMSA using HEC-1B cell nuclear extract and a fluorescently labeled double stranded oligonucleotide comprising of α3SSE–CSE as a probe in the left panel and α12SSE–CSE as a probe in the right panel. Unlabeled competitor DNAs were added to the reactions as indicated in the top panel. The specific DNA complex and the free probes are indicated by arrows.
Figure 5.
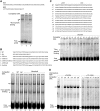
Purification of αSSE–CSE DNA-bound proteins. (A) Schematic diagram of the purification procedure. (B) Binding activity of α6SSE–CSE to the purified fractions was assessed by EMSA using the α6SSE–CSE as a probe. Lane 1, the probe is incubated with SH-SY5Y nuclear extract. Lane 2, with the 800 mM KCl elution fraction of the cation exchange. The flow-through (lanes 3 and 4) and the eluted fractions (lanes 5 and 6) of the affinity purification stage of α6SSE–CSE DNA (α6) and α6SSE–CSE_mut DNA (α6_mut), respectively. (C) Identification of the proteins that bind to the α6SSE–CSE using specific antibodies according to the MS results. Lane 1, the probe is incubated with SH-SY5Y nuclear extract; lanes 2 and 3 EMSAs were carried out in the presence of two different CTCF antibodies, lane 4 YY1 antibody; lane 5 BUB3 antibody. The specific DNA complexes, ‘super-shift’ (SS) complexes and the free probes are indicated by arrows. (D) Western blot analysis of CTCF protein in the flow-through and in the eluted fractions of the affinity purification stage of α6SSE–CSE DNA (α6) and muted DNA (α6_mut).
Figure 6.
Recombinant CTCF binds specifically to several αSSE–CSE DNA sequences. (A) SDS–PAGE analysis of the full-length CTCF and 11ZF CTCF-binding domain proteins, which were synthesized in vitro from the pET-7.1 and pET-11ZF constructs. Luciferase T7 Control DNA no plasmid were used for positive and negative controls, respectively. The in vitro synthesized proteins are fluorescently labeled. Positions of the molecular mass protein markers (on the left) are indicated. The white arrows point to the positions of the in vitro synthesized proteins. (B) EMSAs using _in vitro_-translated luciferase, human CTCF full-length (FL), 11ZF or SH-SY5Y nuclear extract with αSSE–CSE sequences as probes. Lanes 1–3, α12SSE–CSE probe, lanes 4–6, α3SSE–CSE probe and lanes 7–14, α6SSE–CSE probe. The proteins used for binding and the competitor DNA are indicated on the top. The specific complexes between the probe and the recombinant proteins or the nuclear extract are indicated by arrows.
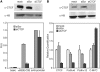
Figure 7.
CTCF is essential for Pcdhα gene-expression. (A) Knocking down CTCF down-regulated the expression of α6 promoter-driven reporter gene. Top panel, western blot analysis with either CTCF or H3 antibodies of SH-SY5Y cells transfected with siRNA-CTCF scrambled siRNA or mock. Bottom panel, 72-h post-siRNA transfction, luciferase reporter gene driven by Pcdhα6, basic and SV40 promoters were transfected into SH-SY5Y cell lines together with RSV-Renilla that served as internal control. Twenty-four hours post-transfection firefly and Renilla luciferase activities were measured. The normalized results are the mean of four independent experiments (±SD). (B) Knocking down CTCF down-regulated the expression of endogenous Pcdhα genes. Top panel, western blot analysis with either CTCF or H3 antibodies of HEC-1B cells transfected with siRNA-CTCF scrambled siRNA or mock. Bottom panel, 120-h post-siRNA transfection, mRNA level was measured using relative quantification for endogenous Pcdha6, Pcdha12 genes and c-Myc gene as a positive control.
Similar articles
- Characterization of a cluster of CTCF-binding sites in a protocadherin regulatory region.
Zhai YN, Xu Q, Guo Y, Wu Q. Zhai YN, et al. Yi Chuan. 2016 Apr;38(4):323-36. doi: 10.16288/j.yczz.16-037. Yi Chuan. 2016. PMID: 27103456 - Identification of the cluster control region for the protocadherin-beta genes located beyond the protocadherin-gamma cluster.
Yokota S, Hirayama T, Hirano K, Kaneko R, Toyoda S, Kawamura Y, Hirabayashi M, Hirabayashi T, Yagi T. Yokota S, et al. J Biol Chem. 2011 Sep 9;286(36):31885-95. doi: 10.1074/jbc.M111.245605. Epub 2011 Jul 19. J Biol Chem. 2011. PMID: 21771796 Free PMC article. - Regulatory elements required for the activation and repression of the protocadherin-alpha gene cluster.
Kehayova P, Monahan K, Chen W, Maniatis T. Kehayova P, et al. Proc Natl Acad Sci U S A. 2011 Oct 11;108(41):17195-200. doi: 10.1073/pnas.1114357108. Epub 2011 Sep 26. Proc Natl Acad Sci U S A. 2011. PMID: 21949399 Free PMC article. - Clustered protocadherins.
Chen WV, Maniatis T. Chen WV, et al. Development. 2013 Aug;140(16):3297-302. doi: 10.1242/dev.090621. Development. 2013. PMID: 23900538 Free PMC article. Review. - Regulation of clustered protocadherin genes in individual neurons.
Hirayama T, Yagi T. Hirayama T, et al. Semin Cell Dev Biol. 2017 Sep;69:122-130. doi: 10.1016/j.semcdb.2017.05.026. Epub 2017 Jun 4. Semin Cell Dev Biol. 2017. PMID: 28591566 Review.
Cited by
- CTCF loss induces giant lamellar bodies in Purkinje cell dendrites.
Hirayama T, Kadooka Y, Tarusawa E, Saitoh S, Nakayama H, Hoshino N, Nakama S, Fukuishi T, Kawanishi Y, Umeshima H, Tomita K, Yoshimura Y, Galjart N, Hashimoto K, Ohno N, Yagi T. Hirayama T, et al. Acta Neuropathol Commun. 2022 Nov 29;10(1):172. doi: 10.1186/s40478-022-01478-6. Acta Neuropathol Commun. 2022. PMID: 36447271 Free PMC article. - CTCF and cohesin help neurons raise their self-awareness.
Dekker J. Dekker J. Proc Natl Acad Sci U S A. 2012 Jun 5;109(23):8799-800. doi: 10.1073/pnas.1206195109. Epub 2012 May 21. Proc Natl Acad Sci U S A. 2012. PMID: 22615386 Free PMC article. No abstract available. - Cohesin subunit Rad21 binds to the HSV-1 genome near CTCF insulator sites during latency in vivo.
Singh P, Neumann DM. Singh P, et al. J Virol. 2021 May 10;95(11):e00364-21. doi: 10.1128/JVI.00364-21. Epub 2021 Mar 10. J Virol. 2021. PMID: 33692212 Free PMC article. - Master regulator genes and their impact on major diseases.
Cai W, Zhou W, Han Z, Lei J, Zhuang J, Zhu P, Wu X, Yuan W. Cai W, et al. PeerJ. 2020 Oct 6;8:e9952. doi: 10.7717/peerj.9952. eCollection 2020. PeerJ. 2020. PMID: 33083114 Free PMC article. - Symmetrical Dose-Dependent DNA-Methylation Profiles in Children with Deletion or Duplication of 7q11.23.
Strong E, Butcher DT, Singhania R, Mervis CB, Morris CA, De Carvalho D, Weksberg R, Osborne LR. Strong E, et al. Am J Hum Genet. 2015 Aug 6;97(2):216-27. doi: 10.1016/j.ajhg.2015.05.019. Epub 2015 Jul 9. Am J Hum Genet. 2015. PMID: 26166478 Free PMC article.
References
- Lomvardas S, Barnea G, Pisapia D, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. - PubMed
- Schmucker D, Clemens J, Shu H, Worby C, Xiao J, Muda M, Dixon J, Zipursky S. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. - PubMed
- Hamada S, Yagi T. The cadherin-related neuronal receptor family: a novel diversified cadherin family at the synapse. Neurosci. Res. 2001;41:207–215. - PubMed