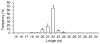Immune-related microRNAs are abundant in breast milk exosomes - PubMed (original) (raw)
Immune-related microRNAs are abundant in breast milk exosomes
Qi Zhou et al. Int J Biol Sci. 2012.
Abstract
Breast milk is a complex liquid rich in immunological components that affect the development of the infant's immune system. Exosomes are membranous vesicles of endocytic origin that are found in various body fluids and that can mediate intercellular communication. MicroRNAs (miRNAs), a well-defined group of non-coding small RNAs, are packaged inside exosomes in human breast milk. Here, we identified 602 unique miRNAs originating from 452 miRNA precursors (pre-miRNAs) in human breast milk exosomes using deep sequencing technology. We found that, out of 87 well-characterized immune-related pre-miRNAs, 59 (67.82%) are presented and enriched in breast milk exosomes (P < 10(-16), χ(2) test). In addition, compared with exogenous synthetic miRNAs, these endogenous immune-related miRNAs are more resistant to relatively harsh conditions. It is, therefore, tempting to speculate that these exosomal miRNAs are transferred from the mother's milk to the infant via the digestive tract, and that they play a critical role in the development of the infant immune system.
Keywords: breast milk; deep sequencing.; exosome; immune-related miRNAs.
Conflict of interest statement
Conflict of Interests: The authors have declared that no conflict of interest exists.
Figures
Figure 1
Length distribution and frequency percent of mappable reads. Values are means ± s.d.
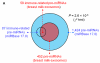
Figure 2
Distribution and characterization of miRNAs in breast milk exosomes. (A) Distribution of pre-miRNAs. Out of 1,424 pre-miRNAs deposited in miRBase 17.0 (blue circle), 87 (6.11%) pre-miRNAs have been designated as immune-related pre-miRNAs based on annotation in the Pathway Central database (SABiosciences, MD, USA). Immune-related pre-miRNAs (59 of 452, 13.05%) are enriched in breast milk exosomes (red circle). _χ_2 test: Number of immune-related miRNAs and others detected in breast milk exosomes compared with the total entries in miRBase 17.0. (B) Top ten most highly expressed unique miRNAs. Plot of the unique miRNAs (starting from the miRNA with the highest counts, x-axis) versus their cumulative % of the total counts from 602 unique miRNAs detected in breast milk exosomes for each small RNA library. Values are means ± s.d. The dashed horizontal line at 62.3% represents the level of the top ten unique miRNAs and the % of individual miRNAs is marked by the gray line. (_C_-E) The expression profile of selected miRNAs in breast milk exosomes. (C) 59 immune-related pre-miRNAs; (D) miR-17-92 cluster and paralogous clusters, and (E) ten well-characterized tissue-specific pre-miRNAs.
Figure 2
Distribution and characterization of miRNAs in breast milk exosomes. (A) Distribution of pre-miRNAs. Out of 1,424 pre-miRNAs deposited in miRBase 17.0 (blue circle), 87 (6.11%) pre-miRNAs have been designated as immune-related pre-miRNAs based on annotation in the Pathway Central database (SABiosciences, MD, USA). Immune-related pre-miRNAs (59 of 452, 13.05%) are enriched in breast milk exosomes (red circle). _χ_2 test: Number of immune-related miRNAs and others detected in breast milk exosomes compared with the total entries in miRBase 17.0. (B) Top ten most highly expressed unique miRNAs. Plot of the unique miRNAs (starting from the miRNA with the highest counts, x-axis) versus their cumulative % of the total counts from 602 unique miRNAs detected in breast milk exosomes for each small RNA library. Values are means ± s.d. The dashed horizontal line at 62.3% represents the level of the top ten unique miRNAs and the % of individual miRNAs is marked by the gray line. (_C_-E) The expression profile of selected miRNAs in breast milk exosomes. (C) 59 immune-related pre-miRNAs; (D) miR-17-92 cluster and paralogous clusters, and (E) ten well-characterized tissue-specific pre-miRNAs.
Figure 2
Distribution and characterization of miRNAs in breast milk exosomes. (A) Distribution of pre-miRNAs. Out of 1,424 pre-miRNAs deposited in miRBase 17.0 (blue circle), 87 (6.11%) pre-miRNAs have been designated as immune-related pre-miRNAs based on annotation in the Pathway Central database (SABiosciences, MD, USA). Immune-related pre-miRNAs (59 of 452, 13.05%) are enriched in breast milk exosomes (red circle). _χ_2 test: Number of immune-related miRNAs and others detected in breast milk exosomes compared with the total entries in miRBase 17.0. (B) Top ten most highly expressed unique miRNAs. Plot of the unique miRNAs (starting from the miRNA with the highest counts, x-axis) versus their cumulative % of the total counts from 602 unique miRNAs detected in breast milk exosomes for each small RNA library. Values are means ± s.d. The dashed horizontal line at 62.3% represents the level of the top ten unique miRNAs and the % of individual miRNAs is marked by the gray line. (_C_-E) The expression profile of selected miRNAs in breast milk exosomes. (C) 59 immune-related pre-miRNAs; (D) miR-17-92 cluster and paralogous clusters, and (E) ten well-characterized tissue-specific pre-miRNAs.
Figure 2
Distribution and characterization of miRNAs in breast milk exosomes. (A) Distribution of pre-miRNAs. Out of 1,424 pre-miRNAs deposited in miRBase 17.0 (blue circle), 87 (6.11%) pre-miRNAs have been designated as immune-related pre-miRNAs based on annotation in the Pathway Central database (SABiosciences, MD, USA). Immune-related pre-miRNAs (59 of 452, 13.05%) are enriched in breast milk exosomes (red circle). _χ_2 test: Number of immune-related miRNAs and others detected in breast milk exosomes compared with the total entries in miRBase 17.0. (B) Top ten most highly expressed unique miRNAs. Plot of the unique miRNAs (starting from the miRNA with the highest counts, x-axis) versus their cumulative % of the total counts from 602 unique miRNAs detected in breast milk exosomes for each small RNA library. Values are means ± s.d. The dashed horizontal line at 62.3% represents the level of the top ten unique miRNAs and the % of individual miRNAs is marked by the gray line. (_C_-E) The expression profile of selected miRNAs in breast milk exosomes. (C) 59 immune-related pre-miRNAs; (D) miR-17-92 cluster and paralogous clusters, and (E) ten well-characterized tissue-specific pre-miRNAs.
Figure 2
Distribution and characterization of miRNAs in breast milk exosomes. (A) Distribution of pre-miRNAs. Out of 1,424 pre-miRNAs deposited in miRBase 17.0 (blue circle), 87 (6.11%) pre-miRNAs have been designated as immune-related pre-miRNAs based on annotation in the Pathway Central database (SABiosciences, MD, USA). Immune-related pre-miRNAs (59 of 452, 13.05%) are enriched in breast milk exosomes (red circle). _χ_2 test: Number of immune-related miRNAs and others detected in breast milk exosomes compared with the total entries in miRBase 17.0. (B) Top ten most highly expressed unique miRNAs. Plot of the unique miRNAs (starting from the miRNA with the highest counts, x-axis) versus their cumulative % of the total counts from 602 unique miRNAs detected in breast milk exosomes for each small RNA library. Values are means ± s.d. The dashed horizontal line at 62.3% represents the level of the top ten unique miRNAs and the % of individual miRNAs is marked by the gray line. (_C_-E) The expression profile of selected miRNAs in breast milk exosomes. (C) 59 immune-related pre-miRNAs; (D) miR-17-92 cluster and paralogous clusters, and (E) ten well-characterized tissue-specific pre-miRNAs.
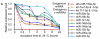
Figure 3
Characterization of breast milk miRNAs. The expression changes of a spiked-in C. elegans miRNA and of the top ten most highly expressed unique miRNAs from the small RNA libraries across various harsh conditions. Total RNA was extracted and then analyzed by q-PCR. Breast milk was (A) incubated at 26°C for 0.5, 1, 2, 4, 8 or 24 hours, (B) subjected to six freeze-thaw cycles at -20°C, (C) treated with RNase A and T1 for 60 min at 37°C, and (D) incubated at 100°C for 10 min.
Figure 3
Characterization of breast milk miRNAs. The expression changes of a spiked-in C. elegans miRNA and of the top ten most highly expressed unique miRNAs from the small RNA libraries across various harsh conditions. Total RNA was extracted and then analyzed by q-PCR. Breast milk was (A) incubated at 26°C for 0.5, 1, 2, 4, 8 or 24 hours, (B) subjected to six freeze-thaw cycles at -20°C, (C) treated with RNase A and T1 for 60 min at 37°C, and (D) incubated at 100°C for 10 min.
Figure 3
Characterization of breast milk miRNAs. The expression changes of a spiked-in C. elegans miRNA and of the top ten most highly expressed unique miRNAs from the small RNA libraries across various harsh conditions. Total RNA was extracted and then analyzed by q-PCR. Breast milk was (A) incubated at 26°C for 0.5, 1, 2, 4, 8 or 24 hours, (B) subjected to six freeze-thaw cycles at -20°C, (C) treated with RNase A and T1 for 60 min at 37°C, and (D) incubated at 100°C for 10 min.
Figure 3
Characterization of breast milk miRNAs. The expression changes of a spiked-in C. elegans miRNA and of the top ten most highly expressed unique miRNAs from the small RNA libraries across various harsh conditions. Total RNA was extracted and then analyzed by q-PCR. Breast milk was (A) incubated at 26°C for 0.5, 1, 2, 4, 8 or 24 hours, (B) subjected to six freeze-thaw cycles at -20°C, (C) treated with RNase A and T1 for 60 min at 37°C, and (D) incubated at 100°C for 10 min.
Similar articles
- Lactation-related microRNA expression profiles of porcine breast milk exosomes.
Gu Y, Li M, Wang T, Liang Y, Zhong Z, Wang X, Zhou Q, Chen L, Lang Q, He Z, Chen X, Gong J, Gao X, Li X, Lv X. Gu Y, et al. PLoS One. 2012;7(8):e43691. doi: 10.1371/journal.pone.0043691. Epub 2012 Aug 24. PLoS One. 2012. PMID: 22937080 Free PMC article. - Exploration of microRNAs in porcine milk exosomes.
Chen T, Xi QY, Ye RS, Cheng X, Qi QE, Wang SB, Shu G, Wang LN, Zhu XT, Jiang QY, Zhang YL. Chen T, et al. BMC Genomics. 2014 Feb 5;15(1):100. doi: 10.1186/1471-2164-15-100. BMC Genomics. 2014. PMID: 24499489 Free PMC article. - Breast Milk-Derived Extracellular Vesicles Enriched in Exosomes From Mothers With Type 1 Diabetes Contain Aberrant Levels of microRNAs.
Mirza AH, Kaur S, Nielsen LB, Størling J, Yarani R, Roursgaard M, Mathiesen ER, Damm P, Svare J, Mortensen HB, Pociot F. Mirza AH, et al. Front Immunol. 2019 Oct 25;10:2543. doi: 10.3389/fimmu.2019.02543. eCollection 2019. Front Immunol. 2019. PMID: 31708933 Free PMC article. - Exosome-Derived MicroRNAs of Human Milk and Their Effects on Infant Health and Development.
Melnik BC, Stremmel W, Weiskirchen R, John SM, Schmitz G. Melnik BC, et al. Biomolecules. 2021 Jun 7;11(6):851. doi: 10.3390/biom11060851. Biomolecules. 2021. PMID: 34200323 Free PMC article. Review. - Xeno-miRNA in Maternal-Infant Immune Crosstalk: An Aid to Disease Alleviation.
Stephen BJ, Pareek N, Saeed M, Kausar MA, Rahman S, Datta M. Stephen BJ, et al. Front Immunol. 2020 Mar 24;11:404. doi: 10.3389/fimmu.2020.00404. eCollection 2020. Front Immunol. 2020. PMID: 32269563 Free PMC article. Review.
Cited by
- Human Breast Milk Exosomes: Affecting Factors, Their Possible Health Outcomes, and Future Directions in Dietetics.
Çelik E, Cemali Ö, Şahin TÖ, Deveci G, Biçer NÇ, Hirfanoğlu İM, Ağagündüz D, Budán F. Çelik E, et al. Nutrients. 2024 Oct 17;16(20):3519. doi: 10.3390/nu16203519. Nutrients. 2024. PMID: 39458514 Free PMC article. Review. - MicroRNA Biogenesis, Gene Regulation Mechanisms, and Availability in Foods.
de Mello AS, Ferguson BS, Shebs-Maurine EL, Giotto FM. de Mello AS, et al. Noncoding RNA. 2024 Oct 11;10(5):52. doi: 10.3390/ncrna10050052. Noncoding RNA. 2024. PMID: 39452838 Free PMC article. Review. - Pig Milk Exosome Packaging ssc-miR-22-3p Alleviates Pig Intestinal Epithelial Cell Injury and Inflammatory Response by Targeting MAPK14.
Li J, Hu H, Fu P, Yang Q, Wang P, Gao X, Yang J, Gun S, Huang X. Li J, et al. Int J Mol Sci. 2024 Oct 5;25(19):10715. doi: 10.3390/ijms251910715. Int J Mol Sci. 2024. PMID: 39409044 Free PMC article. - Relationship of MicroRNA according to Immune Components of Breast Milk in Korean Lactating Mothers.
Choi YJ, Lee DH, Song J, Kim KU, Min H, Chung SH, Kim TH, Kim CY, Kang I, Lee NM, Yi DY. Choi YJ, et al. Pediatr Gastroenterol Hepatol Nutr. 2024 Sep;27(5):322-331. doi: 10.5223/pghn.2024.27.5.322. Epub 2024 Sep 9. Pediatr Gastroenterol Hepatol Nutr. 2024. PMID: 39319280 Free PMC article. - Alternatives of mesenchymal stem cell-derived exosomes as potential therapeutic platforms.
Lee S, Jung SY, Yoo D, Go D, Park JY, Lee JM, Um W. Lee S, et al. Front Bioeng Biotechnol. 2024 Sep 9;12:1478517. doi: 10.3389/fbioe.2024.1478517. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 39315312 Free PMC article. Review.
References
- Admyre C, Johansson SM, Qazi KR, Filen JJ, Lahesmaa R. et al. Exosomes with immune modulatory features are present in human breast milk. J Immunol. 2007;179:1969–1978. - PubMed
- Petherick A. Development: mother's milk: a rich opportunity. Nature. 2010;468:S5–7. - PubMed
- Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. - PubMed
- Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–1920. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources