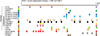Identification of RNA-protein interaction networks using PAR-CLIP - PubMed (original) (raw)
Review
. 2012 Mar-Apr;3(2):159-77.
doi: 10.1002/wrna.1103. Epub 2011 Dec 27.
Affiliations
- PMID: 22213601
- PMCID: PMC3711140
- DOI: 10.1002/wrna.1103
Review
Identification of RNA-protein interaction networks using PAR-CLIP
Manuel Ascano et al. Wiley Interdiscip Rev RNA. 2012 Mar-Apr.
Abstract
All mRNA molecules are subject to some degree of post-transcriptional gene regulation (PTGR) involving sequence-dependent modulation of splicing, cleavage and polyadenylation, editing, transport, stability, and translation. The recent introduction of deep-sequencing technologies enabled the development of new methods for broadly mapping interaction sites between RNA-binding proteins (RBPs) and their RNA target sites. In this article, we review crosslinking and immunoprecipitation (CLIP) methods adapted for large-scale identification of target RNA-binding sites and the respective RNA recognition elements. CLIP methods have the potential to detect hundreds of thousands of binding sites in single experiments although the separation of signal from noise can be challenging. As a consequence, each CLIP method has developed different strategies to distinguish true targets from background. We focus on photoactivatable ribonucleoside-enhanced CLIP, which relies on the intracellular incorporation of photoactivatable ribonucleoside analogs into nascent transcripts, and yields characteristic sequence changes upon crosslinking that facilitate the separation of signal from noise. The precise knowledge of the position and distribution of binding sites across mature and primary mRNA transcripts allows critical insights into cellular localization and regulatory function of the examined RBP. When coupled with other systems-wide approaches measuring transcript and protein abundance, the generation of high-resolution RBP-binding site maps across the transcriptome will broaden our understanding of PTGR and thereby lead to new strategies for therapeutic treatment of genetic diseases perturbing these processes.
Copyright © 2011 John Wiley & Sons, Ltd.
Figures
FIGURE 1
RNA-binding proteins (RBPs) and their RNA-binding domains (RBDs). (a) The most frequently occurring RBDs present in approximately 400 human proteins are shown. Domain names are according to Pfam nomenclature. RBPs containing more than one type of RBD are counted multiple times. (b) The domain composition of RBPs is further resolved. The top panel indicates single versus repeated occurrence of the same RBD in RBPs. The middle panel resolves combinations of RBDs within RBPs, and the bottom panel indicates how many RBPs with at least one member of RBD are combined with another enzymatically active protein domain such as nuclease, helicase, or with protein–protein interaction domains. RNases and helicases without auxiliary RBDs were excluded in the bottom panel.
FIGURE 2
Outline of photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) and other CLIP methodologies. (a) High-throughput sequencing CLIP (HiTS-CLIP, left panel), individual nucleotide resolution CLIP (iCLIP, middle panel), and individual nucleotide resolution crosslinking affinity purification (iCLAP, right panel) utilize short-wavelength ultraviolet (UV) light of 254 nm (UV-254) to crosslink RNA to interacting RNA-binding proteins (RBPs) in living cells or fresh tissue prior to lysis. (b) PAR-CLIP first incorporates photoactivatable thioribonucleosides into nascent transcripts and then crosslinks by using long-wavelength UV-365. Isolation of RNA–RBP complexes is achieved either by immunoprecipitation (IP) (PAR-CLIP, HiTS-CLIP, and iCLIP) or by streptavidin/IMAC double affinity purification (iCLAP). Similar to iCLAP, the crosslinking and analysis of cDNAs (CRAC) method (not shown) utilizes double affinity enrichment of an RBP in which the second purification is performed under denaturing conditions. The addition of 3′ and 5′ adapters differ based on the CLIP method. iCLIP and iCLAP methods circularize an ssDNA intermediate to capture sequences generated from prematurely terminated products of reverse transcription (RT) at the crosslinking sites. In PAR-CLIP, RT through a thioribonucleoside crosslinked RNA, followed by polymerized chain reaction (PCR) amplification, leads to a characteristic mutation [T to C when using 4-thiouridine (4SU) and G to A when using 6-thioguanosine (6SG)]. Scoring for these mutations leads to the differentiation of clusters of crosslinked reads from clusters originating from non-crosslinked background sequences including abundant cellular RNAs.
FIGURE 3
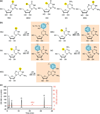
Photoactivatable ribonucleoside analogs. (a) Halogenated and thione-containing ribonucleoside analogs have been used for photocrosslinking to probe RNA–RNA as well as RNA–protein interactions. (b) Predicted structures of ribonucleoside analogs upon ultraviolet (UV) 365 crosslinking to aromatic amino acid side chains. The structure upon crosslinking to tyrosine is shown. (c) Analysis of the ratio of 4-thiouridine (4SU) incorporation into cellular RNAs. Total RNA (0.2 OD260 units) recovered from HEK293 cells cultured in medium supplemented with 0.1 mM 4SU for 16 h, was digested to mononucleosides using snake venom phosphodiesterase and alkaline phosphatase and analyzed by reverse-phase high-performance liquid chromatography (HPLC). The peaks corresponding to C, U, G, and A were detected at 280 nm and the peak corresponding to 4SU is detected at 330 nm. An incorporation rate of 4% 4SU was calculated based on the absorption coefficients for U and 4SU. 4SU substitution rates of 1–4% were also observed with other cell lines. 5BrU, 5-bromouridine; 5IU, 5-iodouridine; 5IC, 5-iodocytidine; 6SG, 6-thioguanosine.
FIGURE 4
Photoactivated crosslinking of thioribonucleosides. (a) Photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) of HEK293 cells expressing FLAG/hemagglutinin (HA)-tagged IGF2BP2 containing 4 RRM and 2 K homology (KH) RNA-binding domains (RBDs). The top panel shows the autoradiograph of radiolabeled RNA crosslinked to IGF2BP2 and resolved after immunoprecipitation (IP) by protein-denaturing sodium dodecyl sulfate (SDS) gel electrophoresis. The expected molecular weight of the epitope tagged IGF2BP2 is approximately 75 kDa. The bottom panel shows an anti-HA immunoblot (IB) to control for the amount of tagged IGF2BP2 protein loaded on the SDS gel. (b) Top panel: ultraviolet (UV) 254 crosslinking products of uridine and aromatic side chain amino acids (Tyr, Phe, and Trp). Reverse transcriptase typically stalls at the crosslinked adduct, but can occasionally skip the lesion resulting in a deletion within the cDNA corresponding to the crosslinked position. The bottom panel shows UV-365 crosslinking product for 4-thiouridine (4SU) to aromatic side chain amino acids. The structure of the 4SU crosslink product is distinct from the UV-254 crosslinked photoadduct. Reverse transcriptase also stalls at the lesion, but when it reads through it preferably incorporates dG directly opposite the photoadduct, leading to the characteristic T to C transitions defining the sites of 4SU crosslinking.
FIGURE 5
Examples of photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) clusters of overlapping sequence reads and the corresponding RNA recognition elements (RREs). Clusters of overlapping sequence reads obtained from PAR-CLIP of the RNA-binding proteins (RBPs) PUM2, QKI, and IGF2BP family members. Reads were aligned to the reference sequence of the indicated gene transcripts; only the most frequent reads are shown. The sequence read copy numbers and the number of mismatches to the reference sequence are indicated to the right; they all represent T to C mutations (shown in red letters). Blue boxes indicate the location(s) of the RREs, as defined by Phylo-Gibbs motif analysis (right panels).
FIGURE 6
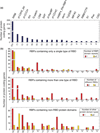
Photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) RNA-binding proteins (RBPs) binding sites for CDK1 mRNA. The CDK1 mRNA is 1923 nt long (NM_001786.4). This figure illustrates only the exonic binding sites identified for several nuclear and cytoplasmic-localized RBPs. The subcellular localization of each RBP is shown. The reported subcellular distribution of some RBPs would indicate that they are predominantly nuclear (FUS), versus cytosplamic (PUM2). However, the nuclear to cytoplasmic ratios of RBPs relative to each other are not known, nor where they would associate with their mRNA targets.
Similar articles
- PAR-CliP--a method to identify transcriptome-wide the binding sites of RNA binding proteins.
Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. Hafner M, et al. J Vis Exp. 2010 Jul 2;(41):2034. doi: 10.3791/2034. J Vis Exp. 2010. PMID: 20644507 Free PMC article. - PAR-CLIP: A Method for Transcriptome-Wide Identification of RNA Binding Protein Interaction Sites.
Danan C, Manickavel S, Hafner M. Danan C, et al. Methods Mol Biol. 2022;2404:167-188. doi: 10.1007/978-1-0716-1851-6_9. Methods Mol Biol. 2022. PMID: 34694609 - Transcriptome-wide Identification of RNA-binding Protein Binding Sites Using Photoactivatable-Ribonucleoside-Enhanced Crosslinking Immunoprecipitation (PAR-CLIP).
Maatz H, Kolinski M, Hubner N, Landthaler M. Maatz H, et al. Curr Protoc Mol Biol. 2017 Apr 3;118:27.6.1-27.6.19. doi: 10.1002/cpmb.35. Curr Protoc Mol Biol. 2017. PMID: 28369676 - [Advances in technologies for large-scale enrichment and identification of ribonucleic acid-protein complexes].
Fan Z, Qin W. Fan Z, et al. Se Pu. 2021 Feb;39(2):105-111. doi: 10.3724/SP.J.1123.2020.07019. Se Pu. 2021. PMID: 34227341 Free PMC article. Review. Chinese. - Optimization of PAR-CLIP for transcriptome-wide identification of binding sites of RNA-binding proteins.
Garzia A, Meyer C, Morozov P, Sajek M, Tuschl T. Garzia A, et al. Methods. 2017 Apr 15;118-119:24-40. doi: 10.1016/j.ymeth.2016.10.007. Epub 2016 Oct 17. Methods. 2017. PMID: 27765618 Free PMC article. Review.
Cited by
- System-wide identification of RNA-binding proteins by interactome capture.
Castello A, Horos R, Strein C, Fischer B, Eichelbaum K, Steinmetz LM, Krijgsveld J, Hentze MW. Castello A, et al. Nat Protoc. 2013 Mar;8(3):491-500. doi: 10.1038/nprot.2013.020. Epub 2013 Feb 14. Nat Protoc. 2013. PMID: 23411631 - ELAVL1 primarily couples mRNA stability with the 3' UTRs of interferon-stimulated genes.
Rothamel K, Arcos S, Kim B, Reasoner C, Lisy S, Mukherjee N, Ascano M. Rothamel K, et al. Cell Rep. 2021 May 25;35(8):109178. doi: 10.1016/j.celrep.2021.109178. Cell Rep. 2021. PMID: 34038724 Free PMC article. - Augmented noncanonical BMP type II receptor signaling mediates the synaptic abnormality of fragile X syndrome.
Kashima R, Roy S, Ascano M, Martinez-Cerdeno V, Ariza-Torres J, Kim S, Louie J, Lu Y, Leyton P, Bloch KD, Kornberg TB, Hagerman PJ, Hagerman R, Lagna G, Hata A. Kashima R, et al. Sci Signal. 2016 Jun 7;9(431):ra58. doi: 10.1126/scisignal.aaf6060. Sci Signal. 2016. PMID: 27273096 Free PMC article. - Unraveling 3'-end RNA uridylation at nucleotide resolution.
Pirouz M, Ebrahimi AG, Gregory RI. Pirouz M, et al. Methods. 2019 Feb 15;155:10-19. doi: 10.1016/j.ymeth.2018.10.024. Epub 2018 Nov 3. Methods. 2019. PMID: 30395968 Free PMC article. Review. - A versatile assay for RNA-binding proteins in living cells.
Strein C, Alleaume AM, Rothbauer U, Hentze MW, Castello A. Strein C, et al. RNA. 2014 May;20(5):721-31. doi: 10.1261/rna.043562.113. Epub 2014 Mar 24. RNA. 2014. PMID: 24664470 Free PMC article.
References
- Mackay JP, Font J, Segal DJ. The prospects for designer single-stranded RNA-binding proteins. Nat Struct Mol Biol. 2011;18:256–261. - PubMed
- Sonnhammer EL, Eddy SR, Durbin R. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins. 1997;28:405–420. - PubMed
- Timchenko LT, Timchenko NA, Caskey CT, Roberts R. Novel proteins with binding specificity for DNA CTG repeats and RNA Cug repeats: implications for myotonic dystrophy. Hum Mol Genet. 1996;5:115–121. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials