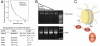Bivalent recognition of nucleosomes by the tandem PHD fingers of the CHD4 ATPase is required for CHD4-mediated repression - PubMed (original) (raw)
Bivalent recognition of nucleosomes by the tandem PHD fingers of the CHD4 ATPase is required for CHD4-mediated repression
Catherine A Musselman et al. Proc Natl Acad Sci U S A. 2012.
Abstract
CHD4 is a catalytic subunit of the NuRD (nucleosome remodeling and deacetylase) complex essential in transcriptional regulation, chromatin assembly and DNA damage repair. CHD4 contains tandem plant homeodomain (PHD) fingers connected by a short linker, the biological function of which remains unclear. Here we explore the combinatorial action of the CHD4 PHD1/2 fingers and detail the molecular basis for their association with chromatin. We found that PHD1/2 targets nucleosomes in a multivalent manner, concomitantly engaging two histone H3 tails. This robust synergistic interaction displaces HP1γ from pericentric sites, inducing changes in chromatin structure and leading to the dispersion of the heterochromatic mark H3K9me3. We demonstrate that recognition of the histone H3 tails by the PHD fingers is required for repressive activity of the CHD4/NuRD complex. Together, our data elucidate the molecular mechanism of multivalent association of the PHD fingers with chromatin and reveal their critical role in the regulation of CHD4 functions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
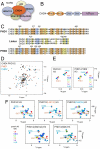
The tandem PHD fingers of CHD4 possess individual histone-binding activities. (A) The CHD4/NuRD complex. (B) CHD4 contains two N-terminal PHD fingers (orange), two chromodomains (blue) and a C-terminal ATPase module (purple). (C) Alignment of the CHD3-CHD5 PHD1/2 sequences. Residues that are strongly, moderately, and weakly conserved in PHD1 and PHD2 fingers are highlighted in orange, blue, and yellow, respectively. (D) An overlay of the 1H,15N HSQC spectra of unbound PHD1/2 (black) and the individual PHD1 (red) and PHD2 (blue) fingers. (E) An overlay of 1H,15N HSQC spectra of the tandem PHD1/2 construct (Left) and PHD1 and PHD2 separately (Right) as increasing amounts of the histone H3 tail peptide (–12) is titrated in. Spectra are color coded according to the molar ratio of protein:peptide. Resonances of the PHD1 domain are labeled in brown and those for the PHD2 domain are labeled in blue. (F) Overlays of 1H,15N HSQC spectra of PHD1/2 (Top Left) and mutants PHD1/2(L381W) (Top Middle), PHD1/2(C460W) (Top Right), PHD1/2(W481A) (Bottom Left), and PHD1/2 (W481A/E439A/E440A/E441A) (Bottom Right) as the histone H3 tail peptide (–12) is titrated in.
Fig. 2.
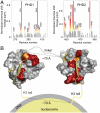
The interdomain organization of PHD1/2 results in a perfect fit for binding to two histone H3 tails in a single nucleosome. (A) The histograms show normalized 1H, 15N chemical shift changes in backbone amides of the PHD1 (Left) and PHD2 (Right) domains of PHD1/2 upon addition of the H3 peptide as a function of residue. Shifts greater than the average plus one-quarter (yellow), one-half (orange) and one (red) standard deviation are highlighted and the residues labeled. These changes are mapped onto the surface of the unbound PHD1 and H3K9me3-bound PHD2 structures in B. (B) A model of the association of CHD4 PHD1/2 with the nucleosome. The PHD1-H3 peptide complex, the linker and orientation with respect to the nucleosome were modeled.
Fig. 3.
PHD1/2 associates bivalently with a single nucleosome. (A) A representative tryptophan fluorescence curve and the two-site fit for PHD1/2 as the tetrasome particle is titrated in. The table shows K d values of the linked PHD1/2 fingers and individual PHD1 and PHD2 for the tetrasome and the unmodified H3 tail peptides, residues 1–12 (averaged over three experiments). (*) Taken from ref. . (B) The tetrasome particle free and in the presence of increasing amounts of PHD1/2 was run on a nondenaturing acrylamide gel and detected by ethidium bromide staining. The tetrasome bands (Top) and the free DNA bands (Bottom) are shown. (C) A model of the concomitant interaction of both PHD modules with the two histone H3 tails.
Fig. 4.
Histone modifications modulate the interaction of PHD1/2. (A) Overlays of 1H, 15N HSQC spectra of PHD1/2 as peptides corresponding to unmodified H3, H3K9ac, H3K9me3, H3K4me3 or H4K20me3 (from left to right) are titrated in. Spectra are color coded according to the protein:peptide molar ratio. (B) Pull-down assays of GST-PHD1/2 with different singly modified histone tail peptides. (C) CHD4 PHD1/2 disrupts pericentric heterochromatin. Representative images of immunofluorescence performed in sorted GFP and GFP-CHD4 PHD1/2 (wild type and the indicated mutants) cells. Cells were spotted onto slides and stained for H3K9me3 and HP1γ. Scale bar, 10 μm. (D) A model of how intranucleosomal binding of CHD4 PHD1/2 can modulate the internucleosomal interaction of HP1γ. (E) Western blot analysis of H3K9me3, H4K20me3, HP1γ, and total H3 acetylation 48 hr after transfection with either GFP or GFP-CHD4 PHD1/2. Total histone H3 (H3 general) was used as a loading control.
Fig. 5.
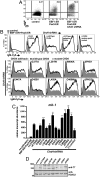
PHD fingers of CHD4 are necessary for transcriptional repressive functions in vivo. (A) Increased display of membrane-bound IgM (mIgM) in response to activated transcription factors (EBF1:ER, Pax5:ER) and depletion of CHD4·μM.2 plasmacytoma cells do not express mIgM in the absence of transcriptional activators (Left). When activated by 4-hydroxytamoxifen (4-OHT), the transcription factors EBF1 (as EBF1:ER) and Pax5 (as Pax5:ER) synergistically activate endogenous Cd79a genes, which enables the assembly of mIgM on the B cell surface at low levels (Middle). shRNA-mediated knockdown of CHD4 greatly increases Cd79a transcription (shown in C) and the display of mIgM in response to EBF1 and Pax5 (Right). Empty retroviruses and luciferase shRNA were used as controls. (B) Effect of mutations in PHD1 and PHD2 in CHD4 on mIgM expression in μM.2 cells. Expression of wild-type CHD4 restores repressive functions when endogenous CHD4 is depleted by 3′UTR-specific shRNA. (Top Row, First Panel) Background mIgM in the presence of EBF1:ER and Pax5:ER. Depletion of CHD4 greatly enhances mIgM (Top Row, Second Panel). Add-back of wild type CHD4 (Top Row, Third Panel) or mutant CHD4 (Top Row, Fourth and Fifth Panels and Middle and Bottom Rows). (C) Relative abundance of mb-1 transcripts from sorted cells in B. CHD4 R1034A (positive control) did not reduce CHD4 activity, while CHD4 K757R (negative control) is inactive. All conditions have activated EBF1:ER and Pax5:ER except control (no activators). Asterisks indicate statistical significance relative to wild type: **= p < 0.001, ***= p < 0.0001. (D) Western blot of CHD4 mutants. Whole cell extracts were prepared from μM.2 cells transduced with CHD4 or control (empty vector) retroviruses and analyzed by blotting with anti-T7 epitope antibodies. HDAC2 served as a nuclear protein loading control.
Similar articles
- How does CHD4 slide nucleosomes?
Reid XJ, Zhong Y, Mackay JP. Reid XJ, et al. Biochem Soc Trans. 2024 Oct 30;52(5):1995-2008. doi: 10.1042/BST20230070. Biochem Soc Trans. 2024. PMID: 39221830 Free PMC article. Review. - Plant homeodomain (PHD) fingers of CHD4 are histone H3-binding modules with preference for unmodified H3K4 and methylated H3K9.
Mansfield RE, Musselman CA, Kwan AH, Oliver SS, Garske AL, Davrazou F, Denu JM, Kutateladze TG, Mackay JP. Mansfield RE, et al. J Biol Chem. 2011 Apr 1;286(13):11779-91. doi: 10.1074/jbc.M110.208207. Epub 2011 Jan 28. J Biol Chem. 2011. PMID: 21278251 Free PMC article. - Inhibition of histone binding by supramolecular hosts.
Allen HF, Daze KD, Shimbo T, Lai A, Musselman CA, Sims JK, Wade PA, Hof F, Kutateladze TG. Allen HF, et al. Biochem J. 2014 May 1;459(3):505-12. doi: 10.1042/BJ20140145. Biochem J. 2014. PMID: 24576085 Free PMC article. - The PHD and chromo domains regulate the ATPase activity of the human chromatin remodeler CHD4.
Watson AA, Mahajan P, Mertens HD, Deery MJ, Zhang W, Pham P, Du X, Bartke T, Zhang W, Edlich C, Berridge G, Chen Y, Burgess-Brown NA, Kouzarides T, Wiechens N, Owen-Hughes T, Svergun DI, Gileadi O, Laue ED. Watson AA, et al. J Mol Biol. 2012 Sep 7;422(1):3-17. doi: 10.1016/j.jmb.2012.04.031. Epub 2012 May 7. J Mol Biol. 2012. PMID: 22575888 Free PMC article. - CHD4 in the DNA-damage response and cell cycle progression: not so NuRDy now.
O'Shaughnessy A, Hendrich B. O'Shaughnessy A, et al. Biochem Soc Trans. 2013 Jun;41(3):777-82. doi: 10.1042/BST20130027. Biochem Soc Trans. 2013. PMID: 23697937 Free PMC article. Review.
Cited by
- How does CHD4 slide nucleosomes?
Reid XJ, Zhong Y, Mackay JP. Reid XJ, et al. Biochem Soc Trans. 2024 Oct 30;52(5):1995-2008. doi: 10.1042/BST20230070. Biochem Soc Trans. 2024. PMID: 39221830 Free PMC article. Review. - ISWI chromatin remodeling complexes recruit NSD2 and H3K36me2 in pericentromeric heterochromatin.
Goto N, Suke K, Yonezawa N, Nishihara H, Handa T, Sato Y, Kujirai T, Kurumizaka H, Yamagata K, Kimura H. Goto N, et al. J Cell Biol. 2024 Aug 5;223(8):e202310084. doi: 10.1083/jcb.202310084. Epub 2024 May 6. J Cell Biol. 2024. PMID: 38709169 Free PMC article. - Chromodomain helicase DNA binding protein 4 in cell fate decisions.
Laureano A, Kim J, Martinez E, Kwan KY. Laureano A, et al. Hear Res. 2023 Sep 1;436:108813. doi: 10.1016/j.heares.2023.108813. Epub 2023 May 30. Hear Res. 2023. PMID: 37329862 Free PMC article. Review. - Gene silencing dynamics are modulated by transiently active regulatory elements.
Vermunt MW, Luan J, Zhang Z, Thrasher AJ, Huang A, Saari MS, Khandros E, Beagrie RA, Zhang S, Vemulamada P, Brilleman M, Lee K, Yano JA, Giardine BM, Keller CA, Hardison RC, Blobel GA. Vermunt MW, et al. Mol Cell. 2023 Mar 2;83(5):715-730.e6. doi: 10.1016/j.molcel.2023.02.006. Mol Cell. 2023. PMID: 36868189 Free PMC article. - DLX1 and the NuRD complex cooperate in enhancer decommissioning and transcriptional repression.
Price JD, Lindtner S, Ypsilanti A, Binyameen F, Johnson JR, Newton BW, Krogan NJ, Rubenstein JLR. Price JD, et al. Development. 2022 Jun 1;149(11):dev199508. doi: 10.1242/dev.199508. Epub 2022 Jun 13. Development. 2022. PMID: 35695185 Free PMC article.
References
- Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. - PubMed
- Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. - PubMed
- Xue Y, et al. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. - PubMed
- Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. - PubMed
- Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–5438. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA113472/CA/NCI NIH HHS/United States
- Z01 ES101765/Intramural NIH HHS/United States
- R01 AI081878/AI/NIAID NIH HHS/United States
- GM096863/GM/NIGMS NIH HHS/United States
- R01 CA113472/CA/NCI NIH HHS/United States
- T32 AI007405/AI/NIAID NIH HHS/United States
- F32 HL096399/HL/NHLBI NIH HHS/United States
- F32HL096399/HL/NHLBI NIH HHS/United States
- P30 CA046934/CA/NCI NIH HHS/United States
- R01 AI054661/AI/NIAID NIH HHS/United States
- T32 AI07405/AI/NIAID NIH HHS/United States
- R37 GM059785/GM/NIGMS NIH HHS/United States
- R01 GM096863/GM/NIGMS NIH HHS/United States
- AI081878/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases