Identification of surprisingly diverse type IV pili, across a broad range of gram-positive bacteria - PubMed (original) (raw)
Identification of surprisingly diverse type IV pili, across a broad range of gram-positive bacteria
Saheed Imam et al. PLoS One. 2011.
Abstract
Background: In Gram-negative bacteria, type IV pili (TFP) have long been known to play important roles in such diverse biological phenomena as surface adhesion, motility, and DNA transfer, with significant consequences for pathogenicity. More recently it became apparent that Gram-positive bacteria also express type IV pili; however, little is known about the diversity and abundance of these structures in Gram-positives. Computational tools for automated identification of type IV pilins are not currently available.
Results: To assess TFP diversity in Gram-positive bacteria and facilitate pilin identification, we compiled a comprehensive list of putative Gram-positive pilins encoded by operons containing highly conserved pilus biosynthetic genes (pilB, pilC). A surprisingly large number of species were found to contain multiple TFP operons (pil, com and/or tad). The N-terminal sequences of predicted pilins were exploited to develop PilFind, a rule-based algorithm for genome-wide identification of otherwise poorly conserved type IV pilins in any species, regardless of their association with TFP biosynthetic operons (http://signalfind.org). Using PilFind to scan 53 Gram-positive genomes (encoding >187,000 proteins), we identified 286 candidate pilins, including 214 in operons containing TFP biosynthetic genes (TBG+ operons). Although trained on Gram-positive pilins, PilFind identified 55 of 58 manually curated Gram-negative pilins in TBG+ operons, as well as 53 additional pilin candidates in operons lacking biosynthetic genes in ten species (>38,000 proteins), including 27 of 29 experimentally verified pilins. False positive rates appear to be low, as PilFind predicted only four pilin candidates in eleven bacterial species (>13,000 proteins) lacking TFP biosynthetic genes.
Conclusions: We have shown that Gram-positive bacteria contain a highly diverse set of type IV pili. PilFind can be an invaluable tool to study bacterial cellular processes known to involve type IV pilus-like structures. Its use in combination with other currently available computational tools should improve the accuracy of predicting the subcellular localization of bacterial proteins.
© 2011 Imam et al.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Physical maps of TFP loci in Gram-positive bacteria, compared with representative Gram-negative pil/tad loci.
Arrows represent relative orientation of open reading frames (ORFs); genes are annotated based on high confidence BLASTP or Pfam hits against experimentally verified homologs. ORFs of the same color correspond to genes with similar function; white and gray arrows represent undefined ORFs. The Burkholderia pseudomallei pil and P. aeruginosa tad operons were selected as Gram-negative representatives based on their organizational complexity and experimentally verified components. A = tadA; B = tadB (tad operons) or pilB (pil operons); C = tadC (tad operons) or pilC (pil operons); D = tadD (tad operons) or pilD (pil operons); E, G, Z = tadE, tadG and tadZ, respectively; M, N, O, Q, S, T = pilM, pilN, pilO, pilQ, pilS and pilT, respectively; V = tadV (tad operons) or pilV (pil operons); GA, GB, GC, GD, GE, GF and GG = comGA, comGB, comGC, comGD, comGE, comGF and comGG, respectively. * indicates the absence of a prepilin peptidase cleavage site.
Figure 2. Alignment of PilD/TadV peptidases and Flp pilin signal peptides from Gram-negative and Gram-positive bacteria.
(A) ClustalW alignment of PilD and TadV sequences from the Gram-positive bacterium D. reducens, including the N-terminal Dis P Dis domain (purple shading) characteristic of PilD/ComC homologs and the Peptidase A24 domain (light grey shading) found in both PilD/ComC and TadV homologs. Darker shading indicates predicted peptidase active sites, including the two essential aspartates (D). (B) Alignment of Gram-positive and negative pilins highlighting the Flp motif (shading), conserved TadV cleavage site (arrow), glutamate (E) at +5, and tyrosine (Y) at position +6 in Flp pilins (diverged in Flp-like pilins). Hydrophobic stretches (italics) were predicted by Phobius .
Figure 3. Phylogenetic classification of Gram-positive and Gram-Negative TFP operons.
Phlyogenetic tree depicting the relationship between the three groups of TFP identified in Gram-positive and Gram-negative bacteria, based on PilB/ComGA/TadA homologs (using H. volcanii FlaI as an outgroup). Note that pil, com and tad operons form distinct clades, with Gram-negative bacteria grouped into clusters within each clade. Three distinct monophyletic groups can be identified within the tad clade, two of which encompass TFP operons that do not encode Flp pilins (no Flp_1 and no Flp_2), while the other includes all the tad operons encoding Flp pilins (Flp). Gram-negative sequences here highlighted in grey.
Figure 4. Analysis of type IV pilins features in Gram-positive bacteria training sets.
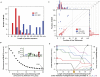
(A) Type IV pilin sequences (red) are shorter than non-type IV pilins (blue). (B) Occurrence of the 6 amino acids core motif for type IV pilin peptidase cleavage, followed by a stretch of uncharged amino acids length 0–20 in 15 genomes (49,601 protein sequences; Table 2) used to define the positive training sets. ∼75% of the these proteins include the six amino acid motif [GAS]-[ACFGILMNPQSTVWY]4-[DE] (inset), and 1381 contain this motif followed by 20 uncharged amino acids, suggesting that many are false positives. (C) Relative position of the type IV pilin motif and first transmembrane domain. Histograms depict the motif position (right) and transmembrane domain position (top) for type IV pilins (red) and non-type IV pilins (blue) in the training sets, only position within 100 amino acids are shown. Note that motif distribution is relatively even in non-type IV pilins, but occurs strictly within the first 35 amino acid residues in type IV pilins. Diagonal dashed lines indicate ±13 amino acids distance between the type IV pilin motif and the first TM domain. In the scatter plot, number indicates the number of TM domains. (D) Assessing the effect of length and amino acid composition of the stretch subsequent to the type IV pilin cleavage pattern. Colored lines indicate the impact of permitting 0, 1, 2, 3, 4 or 5 charged amino acids within the hydrophobic stretch of length 8–20 amino acids. Solid lines represent false positives, and dashed lines false negatives. Seeking a hydrophobic stretch of 14 amino acids with no charged side chains (yellow triangle) yields optimal performance.
Figure 5. Combining features for better identification of type IV pilins.
Criteria applied to the 58 protein positive training dataset, the 116 protein negative training dataset, and 15 Gram-positive bacterial genomes from which the positive training set was assembled; “Short”, protein sequence length ≤350 amino acids; “TM”, presence of a transmembrane domain, a single TM, or the first TM within the N-terminal 50 amino acids; “Motif”, presence of the characteristic prepilin peptidase cleavage recognition site followed by 14 non-charged amino acids, or in close proximity (≤13 amino acids) to the first TM. Green shading indicates criteria applied in these computational experiments. “TP”, true positive; “TN”, true negative; “proteome”, the proteins satisfying the criteria applied.
Figure 6. Comparison of type IV pilin predictions using Hidden Markov Model (HMM)- and regular-expression (RE)-based approaches.
Venn diagram indicating the total number of type IV pilins predicted in 48 genomes (38 Gram-positive and 10 Gram-negative) using an HMM, a RE, or manual curation. The HMM approach displays high specificity, but is only capable of identifying a relatively small subset of the curated type IV pilins (106 of 218), and 24 new pilin candidates. The RE-based approach identified a much larger number of curated type IV pilin (210 of 218), along with 110 new pilin candidates, including all those of identified by the HMM-based approach, respectively. Numbers in parenthesis indicate data for the True Positive “+”, and False Negatives “−”.
Similar articles
- Global biochemical and structural analysis of the type IV pilus from the Gram-positive bacterium Streptococcus sanguinis.
Berry JL, Gurung I, Anonsen JH, Spielman I, Harper E, Hall AMJ, Goosens VJ, Raynaud C, Koomey M, Biais N, Matthews S, Pelicic V. Berry JL, et al. J Biol Chem. 2019 Apr 26;294(17):6796-6808. doi: 10.1074/jbc.RA118.006917. Epub 2019 Mar 5. J Biol Chem. 2019. PMID: 30837269 Free PMC article. - Pilins in gram-positive bacteria: A structural perspective.
Krishnan V. Krishnan V. IUBMB Life. 2015 Jul;67(7):533-43. doi: 10.1002/iub.1400. Epub 2015 Jul 14. IUBMB Life. 2015. PMID: 26178080 Review. - Pili in Gram-negative and Gram-positive bacteria - structure, assembly and their role in disease.
Proft T, Baker EN. Proft T, et al. Cell Mol Life Sci. 2009 Feb;66(4):613-35. doi: 10.1007/s00018-008-8477-4. Cell Mol Life Sci. 2009. PMID: 18953686 Free PMC article. Review. - Crystal Structure of the Minor Pilin CofB, the Initiator of CFA/III Pilus Assembly in Enterotoxigenic Escherichia coli.
Kolappan S, Ng D, Yang G, Harn T, Craig L. Kolappan S, et al. J Biol Chem. 2015 Oct 23;290(43):25805-18. doi: 10.1074/jbc.M115.676106. Epub 2015 Aug 31. J Biol Chem. 2015. PMID: 26324721 Free PMC article. - Biological roles of nontypeable Haemophilus influenzae type IV pilus proteins encoded by the pil and com operons.
Carruthers MD, Tracy EN, Dickson AC, Ganser KB, Munson RS Jr, Bakaletz LO. Carruthers MD, et al. J Bacteriol. 2012 Apr;194(8):1927-33. doi: 10.1128/JB.06540-11. Epub 2012 Feb 10. J Bacteriol. 2012. PMID: 22328674 Free PMC article.
Cited by
- Evaluation of genetic diversity among strains of the human gut commensal Bifidobacterium adolescentis.
Duranti S, Milani C, Lugli GA, Mancabelli L, Turroni F, Ferrario C, Mangifesta M, Viappiani A, Sánchez B, Margolles A, van Sinderen D, Ventura M. Duranti S, et al. Sci Rep. 2016 Apr 1;6:23971. doi: 10.1038/srep23971. Sci Rep. 2016. PMID: 27035119 Free PMC article. - SignalP: The Evolution of a Web Server.
Nielsen H, Teufel F, Brunak S, von Heijne G. Nielsen H, et al. Methods Mol Biol. 2024;2836:331-367. doi: 10.1007/978-1-0716-4007-4_17. Methods Mol Biol. 2024. PMID: 38995548 - Draft genome sequence of Staphylococcus hominis strain Hudgins isolated from human skin implicates metabolic versatility and several virulence determinants.
Calkins S, Couger MB, Jackson C, Zandler J, Hudgins GC, Hanafy RA, Budd C, French DP, Hoff WD, Youssef N. Calkins S, et al. Genom Data. 2016 Oct 7;10:91-96. doi: 10.1016/j.gdata.2016.10.003. eCollection 2016 Dec. Genom Data. 2016. PMID: 27766204 Free PMC article. - Monoderm bacteria: the new frontier for type IV pilus biology.
Pelicic V. Pelicic V. Mol Microbiol. 2019 Dec;112(6):1674-1683. doi: 10.1111/mmi.14397. Epub 2019 Oct 8. Mol Microbiol. 2019. PMID: 31556183 Free PMC article. Review. - Surface Display of Small Affinity Proteins on Synechocystis sp. Strain PCC 6803 Mediated by Fusion to the Major Type IV Pilin PilA1.
Cengic I, Uhlén M, Hudson EP. Cengic I, et al. J Bacteriol. 2018 Jul 25;200(16):e00270-18. doi: 10.1128/JB.00270-18. Print 2018 Aug 15. J Bacteriol. 2018. PMID: 29844032 Free PMC article.
References
- Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol. 2004;2:363–378. - PubMed
- Pelicic V. Type IV pili: e pluribus unum? Mol Microbiol. 2008;68:827–837. - PubMed
- Strom MS, Lory S. Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol. 1993;47:565–596. - PubMed
- Albers SV, Pohlschroder M. Diversity of archaeal type IV pilin-like structures. Extremophiles. 2009;13:403–410. - PubMed
- Pohlschroder M, Ghosh A, Tripepi M, Albers SV. Archaeal type IV pilus-like structures-evolutionarily conserved prokaryotic surface organelles. Curr Opin Microbiol. 2011;14:357–363. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous