Selective small molecule inhibition of poly(ADP-ribose) glycohydrolase (PARG) - PubMed (original) (raw)
. 2012 Mar 16;7(3):563-70.
doi: 10.1021/cb200506t. Epub 2012 Jan 26.
Affiliations
- PMID: 22220926
- PMCID: PMC3306470
- DOI: 10.1021/cb200506t
Selective small molecule inhibition of poly(ADP-ribose) glycohydrolase (PARG)
Kristin E Finch et al. ACS Chem Biol. 2012.
Abstract
The poly(ADP-ribose) (PAR) post-translational modification is essential for diverse cellular functions, including regulation of transcription, response to DNA damage, and mitosis. Cellular PAR is predominantly synthesized by the enzyme poly(ADP-ribose) polymerase-1 (PARP-1). PARP-1 is a critical node in the DNA damage response pathway, and multiple potent PARP-1 inhibitors have been described, some of which show considerable promise in the clinic for the treatment of certain cancers. Cellular PAR is efficiently degraded by poly(ADP-ribose) glycohydrolase (PARG), an enzyme for which no potent, readily accessible, and specific inhibitors exist. Herein we report the discovery of small molecules that effectively inhibit PARG in vitro and in cellular lysates. These potent PARG inhibitors can be produced in two chemical steps from commercial starting materials and have complete specificity for PARG over the other known PAR glycohydrolase (ADP-ribosylhydrolase 3, ARH3) and over PARP-1 and thus will be useful tools for studying the biochemistry of PAR signaling.
Figures
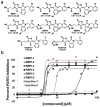
Figure 1
(A) Screening of 224 rhodanine-containing compounds reveals RBPI-1 as a PARG inhibitor, and a collection of >70 compounds was then synthesized based on this scaffold and evaluated for PARG inhibition. From this work RBPI-2, -3, -4, -5, and -6 were identified as potent PARG inhibitors. Inactive-1 and -2 are structurally-related compounds that do not inhibit PARG. (B) Representative dose-response PARG inhibition curves for each compound shown in A. For IC50 determination, compounds were incubated with PARG for 10 min, then 32P-PAR was added and incubated for 2 h at 37°C, at which point the extent of PAR degradation was assessed by separation on TLC plates and phosphorimaging. For TLC images and triplicate IC50 curves please see Supplementary Figure S7.
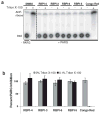
Figure 2
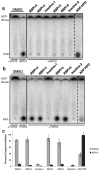
RBPI inhibition of PARG is insensitive to the presence of detergent. Compounds (50 μM) were incubated with PARG in the presence or absence of 0.1% Triton X-100 for 10 min, 32P-PAR was added and incubated for 2 h at 37°C, and the extent of PAR degradation was assessed by separation on TLC plates and phosphorimaging. (A) Representative TLC plate from this experiment; (B) Quantitation of the experiment described above by densitometry, n=3, error bars indicate standard error of the mean.
Figure 3
RBPIs do not inhibit β-lactamase. RBPIs and Congo Red (5-50 μM) were incubated with ß-lactamase (A) in the absence of detergent and (B) in the presence of 0.1% Triton X-100. After addition of the colorimetric substrate, enzyme activity was measured by recording absorbance at 405 nm, n=3, error bars indicate standard error of the mean.
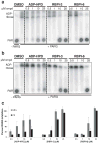
Figure 4
Inhibition of PARG by RBPIs persists even after 2 h incubation with substrate. RBPI-3, -6, and ADP-HPD were incubated with PARG for 10 min, then 32P-PAR was added and incubated at 37°C for (A) 0.5 h or (B) 2 h. The extent of PAR degradation was assessed by separation on TLC plates and phosphorimaging. (C) Quantitation of data shown in A and B, n=3, error bars indicate standard error of the mean.
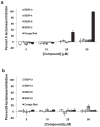
Figure 5
ADP-HPD, but not RBPIs, inhibits ARH3 PAR glycohydrolase activity. Compounds were incubated with (A) ARH3 (256 nM) or (B) PARG (0.24 nM) for 10 min, and then 32P-PAR was added and incubated at 37°C for 2 h. The extent of PAR degradation was visualized by separation on a TLC plate and phosphorimaging. (C) Quantitation of data from part A and B, n=3, error bars indicate standard error of the mean. See Supplementary Figure S10 for IC50 data of ADP-HPD inhibition of ARH3.
Figure 6
Processing of 32P-PAR by cellular lysate (MEF, 1.5 μg) over 60 min in the presence of DMSO, 25 μM RBPI-4, or 25 μM ADP-HPD. Compounds were incubated with lysate for 10 min, then 32P-PAR was added and incubated at 37°C for 0-60 min. The extent of PAR degradation was visualized by separation on a TLC plate and phosphorimaging. Graph is representative of three independent experiments, see Supplementary Figure S13 for triplicate data.
Similar articles
- ADP-ribosylhydrolase 3 (ARH3), not poly(ADP-ribose) glycohydrolase (PARG) isoforms, is responsible for degradation of mitochondrial matrix-associated poly(ADP-ribose).
Niere M, Mashimo M, Agledal L, Dölle C, Kasamatsu A, Kato J, Moss J, Ziegler M. Niere M, et al. J Biol Chem. 2012 May 11;287(20):16088-102. doi: 10.1074/jbc.M112.349183. Epub 2012 Mar 20. J Biol Chem. 2012. PMID: 22433848 Free PMC article. - Discovery and structure-activity relationships of modified salicylanilides as cell permeable inhibitors of poly(ADP-ribose) glycohydrolase (PARG).
Steffen JD, Coyle DL, Damodaran K, Beroza P, Jacobson MK. Steffen JD, et al. J Med Chem. 2011 Aug 11;54(15):5403-13. doi: 10.1021/jm200325s. Epub 2011 Jul 8. J Med Chem. 2011. PMID: 21692479 Free PMC article. - Design and synthesis of phenolic hydrazide hydrazones as potent poly(ADP-ribose) glycohydrolase (PARG) inhibitors.
Islam R, Koizumi F, Kodera Y, Inoue K, Okawara T, Masutani M. Islam R, et al. Bioorg Med Chem Lett. 2014 Aug 15;24(16):3802-6. doi: 10.1016/j.bmcl.2014.06.065. Epub 2014 Jun 27. Bioorg Med Chem Lett. 2014. PMID: 25042255 - Targeting poly(ADP-ribose) glycohydrolase to draw apoptosis codes in cancer.
Tanuma SI, Shibui Y, Oyama T, Uchiumi F, Abe H. Tanuma SI, et al. Biochem Pharmacol. 2019 Sep;167:163-172. doi: 10.1016/j.bcp.2019.06.004. Epub 2019 Jun 6. Biochem Pharmacol. 2019. PMID: 31176615 Review. - New Insights into the Roles of NAD+-Poly(ADP-ribose) Metabolism and Poly(ADP-ribose) Glycohydrolase.
Tanuma S, Sato A, Oyama T, Yoshimori A, Abe H, Uchiumi F. Tanuma S, et al. Curr Protein Pept Sci. 2016;17(7):668-682. doi: 10.2174/1389203717666160419150014. Curr Protein Pept Sci. 2016. PMID: 27817743 Review.
Cited by
- PARP and PARG inhibitors in cancer treatment.
Slade D. Slade D. Genes Dev. 2020 Mar 1;34(5-6):360-394. doi: 10.1101/gad.334516.119. Epub 2020 Feb 6. Genes Dev. 2020. PMID: 32029455 Free PMC article. Review. - Tankyrase-targeted therapeutics: expanding opportunities in the PARP family.
Riffell JL, Lord CJ, Ashworth A. Riffell JL, et al. Nat Rev Drug Discov. 2012 Dec;11(12):923-36. doi: 10.1038/nrd3868. Nat Rev Drug Discov. 2012. PMID: 23197039 Review. - ADP-ribose hydrolases: biological functions and potential therapeutic targets.
Wang J, Wang ZQ, Zong W. Wang J, et al. Expert Rev Mol Med. 2024 Oct 8;26:e21. doi: 10.1017/erm.2024.17. Expert Rev Mol Med. 2024. PMID: 39375922 Free PMC article. Review. - Knockout of PARG110 confers resistance to cGMP-induced toxicity in mammalian photoreceptors.
Sahaboglu A, Tanimoto N, Bolz S, Garrido MG, Ueffing M, Seeliger MW, Löwenheim H, Ekström P, Paquet-Durand F. Sahaboglu A, et al. Cell Death Dis. 2014 May 22;5(5):e1234. doi: 10.1038/cddis.2014.208. Cell Death Dis. 2014. PMID: 24853412 Free PMC article. - Readers of poly(ADP-ribose): designed to be fit for purpose.
Teloni F, Altmeyer M. Teloni F, et al. Nucleic Acids Res. 2016 Feb 18;44(3):993-1006. doi: 10.1093/nar/gkv1383. Epub 2015 Dec 15. Nucleic Acids Res. 2016. PMID: 26673700 Free PMC article. Review.
References
- Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. - PubMed
- d'Adda di Fagagna F, Hande MP, Tong WM, Lansdorp PM, Wang ZQ, Jackson SP. Functions of poly(ADP-ribose) polymerase in controlling telomere length and chromosomal stability. Nat Genet. 1999;23:76–80. - PubMed
- Aguilar-Quesada R, Munoz-Gamez JA, Martin-Oliva D, Peralta-Leal A, Quiles-Perez R, Rodriguez-Vargas JM, Ruiz de Almodovar M, Conde C, Ruiz-Extremera A, Oliver FJ. Modulation of transcription by PARP-1: consequences in carcinogenesis and inflammation. Curr Med Chem. 2007;14:1179–1187. - PubMed
- Davidovic L, Vodenicharov M, Affar EB, Poirier GG. Importance of poly(ADP-ribose) glycohydrolase in the control of poly(ADP-ribose) metabolism. Exp Cell Res. 2001;268:7–13. - PubMed
- Malanga M, Althaus FR. Poly(ADP-ribose) molecules formed during DNA repair in vivo. J Biol Chem. 1994;269:17691–17696. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous