Metabotyping of long-lived mice using 1H NMR spectroscopy - PubMed (original) (raw)
. 2012 Apr 6;11(4):2224-35.
doi: 10.1021/pr2010154. Epub 2012 Feb 27.
Affiliations
- PMID: 22225495
- PMCID: PMC4467904
- DOI: 10.1021/pr2010154
Metabotyping of long-lived mice using 1H NMR spectroscopy
Anisha Wijeyesekera et al. J Proteome Res. 2012.
Abstract
Significant advances in understanding aging have been achieved through studying model organisms with extended healthy lifespans. Employing 1H NMR spectroscopy, we characterized the plasma metabolic phenotype (metabotype) of three long-lived murine models: 30% dietary restricted (DR), insulin receptor substrate 1 null (Irs1-/-), and Ames dwarf (Prop1df/df). A panel of metabolic differences were generated for each model relative to their controls, and subsequently, the three long-lived models were compared to one another. Concentrations of mobile very low density lipoproteins, trimethylamine, and choline were significantly decreased in the plasma of all three models. Metabolites including glucose, choline, glycerophosphocholine, and various lipids were significantly reduced, while acetoacetate, d-3-hydroxybutyrate and trimethylamine-N-oxide levels were increased in DR compared to ad libitum fed controls. Plasma lipids and glycerophosphocholine were also decreased in Irs1-/- mice compared to controls, as were methionine and citrate. In contrast, high density lipoproteins and glycerophosphocholine were increased in Ames dwarf mice, as were methionine and citrate. Pairwise comparisons indicated that differences existed between the metabotypes of the different long-lived mice models. Irs1-/- mice, for example, had elevated glucose, acetate, acetone, and creatine but lower methionine relative to DR mice and Ames dwarfs. Our study identified several potential candidate biomarkers directionally altered across all three models that may be predictive of longevity but also identified differences in the metabolic signatures. This comparative approach suggests that the metabolic networks underlying lifespan extension may not be exactly the same for each model of longevity and is consistent with multifactorial control of the aging process.
Figures
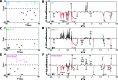
Figure 1
Aliphatic region (δ1H 0.5–4.5) of exemplar 1H NMR plasma spectra. Acquired using (A) standard 1D pulse sequence with presaturation suppression of the water peak, (B) the Carr–Purcell–Meiboom–Gill sequence to attenuate broad signals from proteins and lipoproteins that may overlap signals from low molecular weight metabolites, also using presaturation and (C) a diffusion edited pulse sequence to analyze high molecular weight molecules such as lipids and proteins. *Ethanol contaminant.
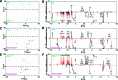
Figure 2
Results of pairwise supervised multivariate modeling performed on CPMG plasma spectroscopic data for each long-lived model vs respective control: O-PLS-DA cross validated scores scatter plot showing the clustering of samples according to metabotype; corresponding O-PLS-DA loadings coefficients plot back-projected with _p_-values, showing the plasma metabolites discriminating between metabotype (models computed using 1 predictive component, 2 orthogonal components in X, 0 orthogonal components in Y, 7-fold cross-validation). (A) O-PLS-DA cross-validated scores of DR and AL mice; (B) O-PLS-DA loadings of DR and AL mice (R2Y = 0.91, Q2Y = 0.85). (C) O-PLS-DA cross-validated scores of Irs1 –/– and WT control mice; (D) O-PLS-DA loadings of Irs1 –/– and WT control mice (R2Y = 0.93, Q2Y = 0.86). (E) O-PLS-DA cross-validated scores of Ames dwarf and WT control mice; (F) O-PLS-DA loadings of Ames dwarf and WT control mice (R2Y = 0.95, Q2Y = 0.84). Refer to Table 1 for assignments of discriminatory metabolites.
Figure 3
Results of pairwise supervised multivariate modeling performed on CPMG plasma spectroscopic data for each long-lived model vs long-lived model (models computed using 1 predictive component, 2 orthogonal components in X, 0 orthogonal components in Y, 7-fold cross-validation). (A) O-PLS-DA cross-validated scores of DR and Irs1 –/– mice; (B) O-PLS-DA loadings of DR and Irs1 –/– mice (R2Y = 0.99, Q2Y = 0.98). (C) O-PLS-DA cross-validated scores of DR and Ames dwarf mice; (D) O-PLS-DA loadings of DR and Ames dwarf mice (R2Y = 0.95, Q2Y = 0.90). (E) O-PLS-DA cross-validated scores of Irs1 –/– and Ames dwarf mice; (F) O-PLS-DA loadings of Irs1 –/– and Ames dwarf (R2Y = 0.99, Q2Y = 0.86). Refer to Table 1 for assignments of discriminatory metabolites.
Figure 4
Results of pairwise supervised multivariate modeling performed on Diffusion Edited plasma spectroscopic data for each long-lived model vs respective control. (A) O-PLS-DA cross-validated scores of DR and AL mice; (B) O-PLS-DA loadings of DR and AL mice (R2 = 0.88, Q2 = 0.85). (C) O-PLS-DA cross-validated scores of Irs1 –/– and WT control mice; (D) O-PLS-DA loadings of Irs1 –/– and WT control mice (R2 = 0.86, Q2 = −0.51). (E) O-PLS-DA cross-validated scores of Ames dwarf and WT control mice; (F) O-PLS-DA loadings of Ames dwarf and WT control mice (R2 = 0.87, Q2 = 0.75). Lipoproteins discriminating between models are labeled in Table 1.
Figure 5
Results of pairwise supervised multivariate modeling performed on Diffusion Edited plasma spectroscopic data for each long-lived model vs long-lived model. (A) O-PLS-DA cross-validated scores of DR and Irs1 –/– mice; (B) O-PLS-DA loadings of DR and Irs1 –/– mice (R2 = 0.89, Q2 = 0.42). (C) O-PLS-DA cross-validated scores of DR and Ames dwarf mice; (D) O-PLS-DA loadings of DR and Ames dwarf mice (R2 = 0.96, Q2 = 0.89). (E) O-PLS-DA cross-validated scores of Irs1 –/– and Ames dwarf mice; (F) O-PLS-DA loadings of Irs1 –/– and Ames dwarf (R2 = 0.93, Q2 = 0.86). Lipoproteins discriminating between models are labeled in Table 1.
Similar articles
- Impact of visceral adipose tissue on longevity and metabolic health: a comparative study of gene expression in perirenal and epididymal fat of Ames dwarf mice.
Zaczek A, Lewiński A, Karbownik-Lewińska M, Lehoczki A, Gesing A. Zaczek A, et al. Geroscience. 2024 Dec;46(6):5925-5938. doi: 10.1007/s11357-024-01131-1. Epub 2024 Mar 22. Geroscience. 2024. PMID: 38517641 Free PMC article. - Abnormalities in metabolic pathways in celiac disease investigated by the metabolic profiling of small intestinal mucosa, blood plasma and urine by NMR spectroscopy.
Upadhyay D, Singh A, Das P, Mehtab J, Dattagupta S, Ahuja V, Makharia GK, Jagannathan NR, Sharma U. Upadhyay D, et al. NMR Biomed. 2020 Aug;33(8):e4305. doi: 10.1002/nbm.4305. Epub 2020 May 11. NMR Biomed. 2020. PMID: 32394522 - Common and unique transcriptional responses to dietary restriction and loss of insulin receptor substrate 1 (IRS1) in mice.
Page MM, Schuster EF, Mudaliar M, Herzyk P, Withers DJ, Selman C. Page MM, et al. Aging (Albany NY). 2018 May 20;10(5):1027-1052. doi: 10.18632/aging.101446. Aging (Albany NY). 2018. PMID: 29779018 Free PMC article. - Translational Metabolomics of Head Injury: Exploring Dysfunctional Cerebral Metabolism with Ex Vivo NMR Spectroscopy-Based Metabolite Quantification.
Wolahan SM, Hirt D, Glenn TC. Wolahan SM, et al. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. PMID: 26269925 Free Books & Documents. Review. - Calorie restriction and dwarf mice in gerontological research.
McKee Alderman J, DePetrillo MA, Gluesenkamp AM, Hartley AC, Verhoff SV, Zavodni KL, Combs TP. McKee Alderman J, et al. Gerontology. 2010;56(4):404-9. doi: 10.1159/000235720. Epub 2009 Aug 19. Gerontology. 2010. PMID: 19690401 Review.
Cited by
- NMR Metabolomics Analysis of Parkinson's Disease.
Lei S, Powers R. Lei S, et al. Curr Metabolomics. 2013;1(3):191-209. doi: 10.2174/2213235X113019990004. Curr Metabolomics. 2013. PMID: 26078917 Free PMC article. - Transcriptomics and Metabonomics Identify Essential Metabolic Signatures in Calorie Restriction (CR) Regulation across Multiple Mouse Strains.
Collino S, Martin FP, Montoliu I, Barger JL, Da Silva L, Prolla TA, Weindruch R, Kochhar S. Collino S, et al. Metabolites. 2013 Oct 11;3(4):881-911. doi: 10.3390/metabo3040881. Metabolites. 2013. PMID: 24958256 Free PMC article. - Landscape of Innovative Methods for Early Diagnosis of Gastric Cancer: A Systematic Review.
Orășeanu A, Brisc MC, Maghiar OA, Popa H, Brisc CM, Șolea SF, Maghiar TA, Brisc C. Orășeanu A, et al. Diagnostics (Basel). 2023 Dec 5;13(24):3608. doi: 10.3390/diagnostics13243608. Diagnostics (Basel). 2023. PMID: 38132192 Free PMC article. Review. - Organization of the Mammalian Metabolome according to Organ Function, Lineage Specialization, and Longevity.
Ma S, Yim SH, Lee SG, Kim EB, Lee SR, Chang KT, Buffenstein R, Lewis KN, Park TJ, Miller RA, Clish CB, Gladyshev VN. Ma S, et al. Cell Metab. 2015 Aug 4;22(2):332-43. doi: 10.1016/j.cmet.2015.07.005. Cell Metab. 2015. PMID: 26244935 Free PMC article. - Gut microbiota: Dietary and social modulation of gut microbiota in the elderly.
Kinross J, Nicholson JK. Kinross J, et al. Nat Rev Gastroenterol Hepatol. 2012 Oct;9(10):563-4. doi: 10.1038/nrgastro.2012.169. Epub 2012 Sep 4. Nat Rev Gastroenterol Hepatol. 2012. PMID: 22945446 No abstract available.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous