Inhibition of microRNA function by antimiR oligonucleotides - PubMed (original) (raw)
Inhibition of microRNA function by antimiR oligonucleotides
Jan Stenvang et al. Silence. 2012.
Abstract
MicroRNAs (miRNAs) have emerged as important post-transcriptional regulators of gene expression in many developmental and cellular processes. Moreover, there is now ample evidence that perturbations in the levels of individual or entire families of miRNAs are strongly associated with the pathogenesis of a wide range of human diseases. Indeed, disease-associated miRNAs represent a new class of targets for the development of miRNA-based therapeutic modalities, which may yield patient benefits unobtainable by other therapeutic approaches. The recent explosion in miRNA research has accelerated the development of several computational and experimental approaches for probing miRNA functions in cell culture and in vivo. In this review, we focus on the use of antisense oligonucleotides (antimiRs) in miRNA inhibition for loss-of-function studies. We provide an overview of the currently employed antisense chemistries and their utility in designing antimiR oligonucleotides. Furthermore, we describe the most commonly used in vivo delivery strategies and discuss different approaches for assessment of miRNA inhibition and potential off-target effects. Finally, we summarize recent progress in antimiR mediated pharmacological inhibition of disease-associated miRNAs, which shows great promise in the development of novel miRNA-based therapeutics.
Figures
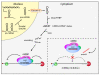
Figure 1
miRNA biogenesis and inhibition of miRNA function by antimiR oligonucleotides. miRNA genes are transcribed by RNA polymerase II into long primary miRNA transcripts, termed pri-miRNAs that are usually several kilobases long and possess a 5' CAP and a poly(A) tail. Pri-miRNAs are processed in the nucleus to ~70 nt pre-miRNAs by the nuclear Microprocessor complex, consisting of DGCR8 and the RNase III enzyme Drosha. Pre-miRNAs are exported to the cytoplasm by Exportin-5 and processed further by Dicer, to ~22 nt double-stranded miRNA duplexes that are loaded into an Argonaute protein in the miRISC and rapidly unwound. During this process the mature miRNA is retained in the miRISC, whereas the complementary strand, known as the miRNA star (miR*) is released. Metazoan miRNAs guide the miRISC to partially complementary sites in the 3' UTRs of target mRNAs to promote their translational repression or deadenylation and degradation. Chemically modified antimiR oligonucleotides sequester the mature miRNA in competition with cellular target mRNAs leading to functional inhibition of the miRNA and derepression of the direct targets.
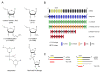
Figure 2
Design of chemically modified antimiR oligonucleotides. (A) Structures of the most commonly used chemical modifications in antimiR oligonucleotides. Locked nucleic acid (LNA) is a bicyclic RNA analogue in which the ribose is locked in a C3'-endo conformation by introduction of a 2'-O,4'-C methylene bridge. The 2'-fluoro (2'-F), 2'-_O_-methoxyethyl (2'-MOE) and 2'-_O_-methyl (2'-_O_-Me) nucleotides are modified at the 2' position of the ribose moiety, whereas a six-membered morpholine ring replaces the sugar moiety in morpholino oligomers. In the phosphorothioate (PS) linkage, sulfur replaces one of the non-bridging oxygen atoms in the phosphate group. (B) Design of chemically modified antimiR oligonucleotides described in this review. (C) Schematic overview of the miRNA inhibition approach using a fully complementary antimiR and a seed-targeting tiny LNA.
Figure 3
Assessment of miRNA inhibition in cultured cells and in vivo. (A) Upper panel. Relative luciferase activity of the miR-21 reporter containing a perfect match miR-21 target site co-transfected into HeLa cells with 1 or 5 nM tiny LNA-antimiR-21, or 5 nM 8-mer 2'-_O_-Me antimiR-21, LNA mismatch (mm) or LNA scramble (scr) control oligonucleotides, respectively. Error bars represent s.e.m. Lower panel. Northern blot analysis of miR-21 in HeLa cells transfected with 5 nM antimiR-21 or LNA scramble control. U6 is shown as control. (B) Relative luciferase activity of the miR-21 reporter co-transfected into HeLa cells, with 5 nM tiny seed-targeting LNAs harbouring single or two adjacent mismatches at all possible nucleotide positions in the antimiR-21 sequence (highlighted in red). (C) Relative luciferase activity of a miR-122 reporter containing a perfect match miR-122 target site co-transfected into HeLa cells with pre-miR-122 and tiny 8-mer antimiR-122 or 15-mer antimiR-122. Error bars represent s.e.m. (D) Northern blot analysis of liver RNAs from mice after treatment with three intravenous doses of 20 mg/kg 8-mer antimiR-122, 15-mer antimiR-122 or LNA scramble control or with saline. The Northern blot was probed for miR-122 and U6. (E) Quantification of the AldoA and Bckdk target mRNAs (same samples as in D, normalized to GAPDH; error bars, s.e.m.; n = 5). (F) Sylamer analyses performed on microarray data from mouse liver RNAs after treatment with three intravenous doses of 20 mg/kg 8-mer antimiR-122 or 15-mer antimiR-122. Shown are Sylamer enrichment landscape plots for 7 nt sequence words. The highlighted words in the plots correspond to canonical miR-122 seed match sites and to perfect match binding sites for the 8-mer antimiR-122. (G) Total plasma cholesterol levels in mice treated with three intravenous injections of 8-mer antimiR-122, 15-mer antimiR-122 or LNA scramble control or with saline (error bars, s.e.m.; n = 5). Adapted from Obad et al. [103].
Similar articles
- Nanoparticle-complexed antimiRs for inhibiting tumor growth and metastasis in prostate carcinoma and melanoma.
Kunz M, Brandl M, Bhattacharya A, Nobereit-Siegel L, Ewe A, Weirauch U, Hering D, Reinert A, Kalwa H, Guzman J, Weigelt K, Wach S, Taubert H, Aigner A. Kunz M, et al. J Nanobiotechnology. 2020 Nov 23;18(1):173. doi: 10.1186/s12951-020-00728-w. J Nanobiotechnology. 2020. PMID: 33228711 Free PMC article. - LNA-mediated microRNA silencing in non-human primates.
Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjärn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. Elmén J, et al. Nature. 2008 Apr 17;452(7189):896-9. doi: 10.1038/nature06783. Epub 2008 Mar 26. Nature. 2008. PMID: 18368051 - Canonical and non-canonical barriers facing antimiR cancer therapeutics.
Cheng CJ, Saltzman WM, Slack FJ. Cheng CJ, et al. Curr Med Chem. 2013;20(29):3582-93. doi: 10.2174/0929867311320290004. Curr Med Chem. 2013. PMID: 23745563 Free PMC article. Review. - The therapeutic potential of microRNAs in cancer.
Thorsen SB, Obad S, Jensen NF, Stenvang J, Kauppinen S. Thorsen SB, et al. Cancer J. 2012 May-Jun;18(3):275-84. doi: 10.1097/PPO.0b013e318258b5d6. Cancer J. 2012. PMID: 22647365 - miRNA therapeutics: a new class of drugs with potential therapeutic applications in the heart.
Bernardo BC, Ooi JY, Lin RC, McMullen JR. Bernardo BC, et al. Future Med Chem. 2015;7(13):1771-92. doi: 10.4155/fmc.15.107. Epub 2015 Sep 24. Future Med Chem. 2015. PMID: 26399457 Review.
Cited by
- MicroRNA-92a-3p Regulates Retinal Angiogenesis by Targeting SGK3 in Vascular Endothelial Cells.
Cui Y, Liu R, Hong Y, Wang Y, Zhu Y, Wen T, Lu J, Mao S, Wang X, Pan J, Luo Y. Cui Y, et al. Invest Ophthalmol Vis Sci. 2022 Oct 3;63(11):19. doi: 10.1167/iovs.63.11.19. Invest Ophthalmol Vis Sci. 2022. PMID: 36269185 Free PMC article. - Potential function of miRNAs in herpetic stromal keratitis.
Mulik S, Bhela S, Rouse BT. Mulik S, et al. Invest Ophthalmol Vis Sci. 2013 Jan 17;54(1):563-73. doi: 10.1167/iovs.12-11094. Invest Ophthalmol Vis Sci. 2013. PMID: 23329734 Free PMC article. Review. - Control of CRK-RAC1 activity by the miR-1/206/133 miRNA family is essential for neuromuscular junction function.
Klockner I, Schutt C, Gerhardt T, Boettger T, Braun T. Klockner I, et al. Nat Commun. 2022 Jun 8;13(1):3180. doi: 10.1038/s41467-022-30778-7. Nat Commun. 2022. PMID: 35676269 Free PMC article. - Highly efficient silencing of microRNA by heteroduplex oligonucleotides.
Yoshioka K, Kunieda T, Asami Y, Guo H, Miyata H, Yoshida-Tanaka K, Sujino Y, Piao W, Kuwahara H, Nishina K, Hara RI, Nagata T, Wada T, Obika S, Yokota T. Yoshioka K, et al. Nucleic Acids Res. 2019 Aug 22;47(14):7321-7332. doi: 10.1093/nar/gkz492. Nucleic Acids Res. 2019. PMID: 31214713 Free PMC article. - Anti-SARS-CoV-2 Strategies and the Potential Role of miRNA in the Assessment of COVID-19 Morbidity, Recurrence, and Therapy.
Narożna M, Rubiś B. Narożna M, et al. Int J Mol Sci. 2021 Aug 12;22(16):8663. doi: 10.3390/ijms22168663. Int J Mol Sci. 2021. PMID: 34445368 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources