Genomic analyses of modern dog breeds - PubMed (original) (raw)
Genomic analyses of modern dog breeds
Heidi G Parker. Mamm Genome. 2012 Feb.
Abstract
A rose may be a rose by any other name, but when you call a dog a poodle it becomes a very different animal than if you call it a bulldog. Both the poodle and the bulldog are examples of dog breeds of which there are >400 recognized worldwide. Breed creation has played a significant role in shaping the modern dog from the length of his leg to the cadence of his bark. The selection and line-breeding required to maintain a breed has also reshaped the genome of the dog, resulting in a unique genetic pattern for each breed. The breed-based population structure combined with extensive morphologic variation and shared human environments have made the dog a popular model for mapping both simple and complex traits and diseases. In order to obtain the most benefit from the dog as a genetic system, it is necessary to understand the effect structured breeding has had on the genome of the species. That is best achieved by looking at genomic analyses of the breeds, their histories, and their relationships to each other.
Figures
Figure 1
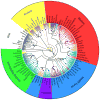
Neighbor joining tree from 85 dog breeds showing the Ancient breed cluster. Ninety-six microsatellite markers were used to calculate the genetic distance between 85 breeds of dog. Using the gray wolf to root the tree, the nine breeds nearest the wolf formed statistically significant branches. The other 76 breeds form a single node indicating shared ancestry and hybridization between the branches. Figure originally published in Science (Parker et al. 2004).
Figure 2
Clustering breeds based on SNP markers increases breed relationship definition. A neighbor joining tree build from comparisons of 10-SNP haplotypes groups 80 dog breeds into approximately 10 breed clusters. Comparable clusters from a microsatellite analysis of 130 breeds (Parker et al. 2007) are indicated by the colored bars outside of the tree. The haplotype tree of breeds was originally published in Nature (Vonholdt et al. 2010).
Figure 3
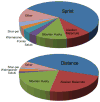
Breed composition of Alaskan Sled dogs. The sled dogs are divided into sprint and distance groups based on their performance level. The dogs that perform as sprinters show a considerable contribution from the Pointer breed that is missing in distance runners. The distance dogs appear to be largely mixtures of classic sled dog breeds. The other group is comprised of 9 breeds with >1% but <2% contribution to at least one group. Data is compiled from Huson et al. (Huson et al. 2010).
Similar articles
- Genetic structure of the purebred domestic dog.
Parker HG, Kim LV, Sutter NB, Carlson S, Lorentzen TD, Malek TB, Johnson GS, DeFrance HB, Ostrander EA, Kruglyak L. Parker HG, et al. Science. 2004 May 21;304(5674):1160-4. doi: 10.1126/science.1097406. Science. 2004. PMID: 15155949 - The effects of dog breed development on genetic diversity and the relative influences of performance and conformation breeding.
Pedersen N, Liu H, Theilen G, Sacks B. Pedersen N, et al. J Anim Breed Genet. 2013 Jun;130(3):236-48. doi: 10.1111/jbg.12017. Epub 2012 Dec 6. J Anim Breed Genet. 2013. PMID: 23679949 - Genetic analysis of the modern Australian labradoodle dog breed reveals an excess of the poodle genome.
Ali MB, Evans JM, Parker HG, Kim J, Pearce-Kelling S, Whitaker DT, Plassais J, Khan QM, Ostrander EA. Ali MB, et al. PLoS Genet. 2020 Sep 10;16(9):e1008956. doi: 10.1371/journal.pgen.1008956. eCollection 2020 Sep. PLoS Genet. 2020. PMID: 32911491 Free PMC article. - Lessons learned from the dog genome.
Wayne RK, Ostrander EA. Wayne RK, et al. Trends Genet. 2007 Nov;23(11):557-67. doi: 10.1016/j.tig.2007.08.013. Epub 2007 Oct 25. Trends Genet. 2007. PMID: 17963975 Review. - The canine genome.
Ostrander EA, Wayne RK. Ostrander EA, et al. Genome Res. 2005 Dec;15(12):1706-16. doi: 10.1101/gr.3736605. Genome Res. 2005. PMID: 16339369 Review.
Cited by
- Comparative analysis of whole exome sequencing kits for the canine genome.
Jang J, Lee YJ, Ko S, Abd El-Aty AM, Gecili I, Jeong JH, Kwon C, Jung TW. Jang J, et al. PLoS One. 2024 Nov 4;19(11):e0312203. doi: 10.1371/journal.pone.0312203. eCollection 2024. PLoS One. 2024. PMID: 39495750 Free PMC article. - Genetic diversity and origin of Kazakh Tobet Dogs.
Perfilyeva A, Bespalova K, Kuzovleva Y, Mussabayev R, Begmanova М, Amirgalyeva A, Vishnyakova O, Nazarenko I, Zhaxsylykova A, Yerzhan A, Perfilyeva Y, Dzhaembaeva T, Khamchukova A, Plakhov K, Torekhanov A, Djansugurova L, Zhunussova G, Bekmanov B. Perfilyeva A, et al. Sci Rep. 2024 Oct 4;14(1):23137. doi: 10.1038/s41598-024-74061-9. Sci Rep. 2024. PMID: 39367220 Free PMC article. - Lymphoma in Border Collies: Genome-Wide Association and Pedigree Analysis.
Soh PXY, Khatkar MS, Williamson P. Soh PXY, et al. Vet Sci. 2023 Sep 19;10(9):581. doi: 10.3390/vetsci10090581. Vet Sci. 2023. PMID: 37756103 Free PMC article. - Identification of Genomic Signatures in Bullmastiff Dogs Using Composite Selection Signals Analysis of 23 Purebred Clades.
Hsu WT, Williamson P, Khatkar MS. Hsu WT, et al. Animals (Basel). 2023 Mar 24;13(7):1149. doi: 10.3390/ani13071149. Animals (Basel). 2023. PMID: 37048405 Free PMC article. - An update on animal models of intervertebral disc degeneration and low back pain: Exploring the potential of artificial intelligence to improve research analysis and development of prospective therapeutics.
Alini M, Diwan AD, Erwin WM, Little CB, Melrose J. Alini M, et al. JOR Spine. 2023 Jan 30;6(1):e1230. doi: 10.1002/jsp2.1230. eCollection 2023 Mar. JOR Spine. 2023. PMID: 36994457 Free PMC article. Review.
References
- Alderton D. Dogs. New York: Dorling Kindersley, Ltd; 2002.
- American Kennel Club. The Complete Dog Book. 19. New York, NY: Howell Book House; 1998. Revised ed.
- American Veterinary Medical Association. US Pet Ownership and Demographics Sourcebook. 1. Schaumburg, ILL: American Veterinary Medical Association; 2002.
- Awano T, Johnson GS, Wade CM, Katz ML, Johnson GC, Taylor JF, Perloski M, Biagi T, Baranowska I, Long S, March PA, Olby NJ, Shelton GD, Khan S, O’Brien DP, Lindblad-Toh K, Coates JR. Genome-wide association analysis reveals a SOD1 mutation in canine degenerative myelopathy that resembles amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2009;106:2794–2799. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous