Chloride intracellular channels modulate acute ethanol behaviors in Drosophila, Caenorhabditis elegans and mice - PubMed (original) (raw)
Chloride intracellular channels modulate acute ethanol behaviors in Drosophila, Caenorhabditis elegans and mice
P Bhandari et al. Genes Brain Behav. 2012 Jun.
Abstract
Identifying genes that influence behavioral responses to alcohol is critical for understanding the molecular basis of alcoholism and ultimately developing therapeutic interventions for the disease. Using an integrated approach that combined the power of the Drosophila, Caenorhabditis elegans and mouse model systems with bioinformatics analyses, we established a novel, conserved role for chloride intracellular channels (CLICs) in alcohol-related behavior. CLIC proteins might have several biochemical functions including intracellular chloride channel activity, modulation of transforming growth factor (TGF)-β signaling, and regulation of ryanodine receptors and A-kinase anchoring proteins. We initially identified vertebrate Clic4 as a candidate ethanol-responsive gene via bioinformatic analysis of data from published microarray studies of mouse and human ethanol-related genes. We confirmed that Clic4 expression was increased by ethanol treatment in mouse prefrontal cortex and also uncovered a correlation between basal expression of Clic4 in prefrontal cortex and the locomotor activating and sedating properties of ethanol across the BXD mouse genetic reference panel. Furthermore, we found that disruption of the sole Clic Drosophila orthologue significantly blunted sensitivity to alcohol in flies, that mutations in two C. elegans Clic orthologues, exc-4 and exl-1, altered behavioral responses to acute ethanol in worms and that viral-mediated overexpression of Clic4 in mouse brain decreased the sedating properties of ethanol. Together, our studies demonstrate key roles for Clic genes in behavioral responses to acute alcohol in Drosophila, C. elegans and mice.
© 2012 The Authors. Genes, Brain and Behavior © 2012 Blackwell Publishing Ltd and International Behavioural and Neural Genetics Society.
Figures
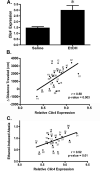
Figure 1. Ethanol-responsive and basal expression of Clic4 in mouse PFC
(A) qRT-PCR analysis of basal (Saline) and ethanol-responsive (EtOH, 4 g/kg, 4 hr) Clic4 expression in DBA2/2J mice. Expression of Clic4 is elevated after ethanol treatment, validating prior microarray results (Kerns et al., 2005) . (B and C) Pearson correlation of Clic4 basal expression in PFC (x-axis) of BXD recombinant inbred lines (numbered points) with ethanol-induced locomotor activity (GeneNetwork trait ID 11962 (Philip et al., 2010)) (B) and initial sensitivity to ethanol-induced rotarod ataxia following first of five injections (onset of ataxia brain ethanol threshold, mg ethanol/g brain – GeneNetwork ID 10144 (Gallaher et al., 1996)) (C). Scattergrams were generated in GeneNetwork (
) using the VCU PFC saline database (Wolen and Miles, unpublished). B6 and D2 strains were not tested in experiments shown in panel B
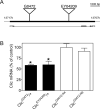
Figure 2. Transposon insertions cause partial loss of function in Drosophila Clic
(A) The Clic locus and transposon insertions. Transcription of Clic is from left to right. The Clic transcription unit is represented by the filled rectangle with nucleotide positions indicated above (coordinates from FlyBase annotation release 5.33). Exons are represented by rectangles below the transcription unit with protein coding sequences and untranslated regions depicted as grey and open rectangles, respectively. Introns are represented as a line and transposons as triangles. Scale bar (upper right) is 1000 bp. Schematic adapted from FlyBase. (B) Whole-body Clic mRNA expression in transposon lines. Expression of Clic mRNA in flies heterozygous for the G0472 and the EY04209 transposons was reduced relative to w 1118 controls.
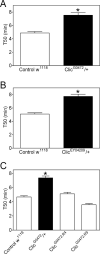
Figure 3. Ethanol sensitivity in Drosophila Clic transposon mutants and revertants
Ethanol sensitivity represented as T50 values in Clic G0472/+ and Clic EY04209/+ (black bars, A and B, respectively) and Control w 1118 flies (open bars). Clic mutants had significantly higher T50 values than controls. (C) Ethanol sensitivity (T50 values) in Clic G0472 heterozygous transposon mutants and revertants. Clic G0472/+ flies (black bar) had higher T50 values than Control w 1118 flies or revertants (Clic G0472.R4 and Clic G0472.R9) (white bars).
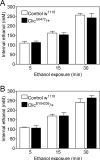
Figure 4. Internal ethanol concentrations in Drosophila Clic mutants
Clic G0472/+ (A) and Clic EY04209/+ (B) were exposed to ethanol vapor for the indicated durations in eRING assays in parallel with Control w 1118 flies. Internal ethanol concentrations were determined as described in Materials and Methods
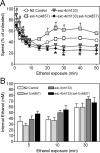
Figure 5. Ethanol sensitivity and acute functional tolerance in C. elegans with mutations in Clic orthologues
(A) Effect of ethanol exposure on relative locomotor speed (percent of untreated animals) in N2 control (open circles) and Clic mutants (exl-1(ok857), light grey squares; exc-4(rh133), dark grey triangles; exc-4(rh133);exl-1(ok857), black diamonds). Data are from 4 independent experiments where 10 worms per genotype contributed to an average speed for a population. (B) Internal ethanol concentrations in N2 control and Clic mutants were determined as described in Materials and Methods
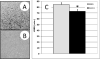
Figure 6. Altered loss of righting reflex (LORR) in mice with AAV2 viral vector-mediated expression of Clic4 in brain
AAV2 vectors expressing a _Clic4_-FLAG fusion protein (AAV-CLIC4; panel A) or empty vector (AAV-IRES; panel B) were stereotactically injected into male DBA/2J mouse PFC. Panels A and B show immunohistochemistry results for FLAG epitope primary antibody staining. Immunohistochemistry was done 2 weeks after the last behavioral studies (~9 weeks after viral injections). Panel C shows that Clic4 over-expressing animals (black) had a shorter duration of LORR following 3.8 g/kg IP of ethanol compared to control (grey). Behavioral testing for LORR was done ~7 weeks after viral injections.
Similar articles
- Chloride intracellular channel 4 (CLIC4) expression profile in the mouse medial prefrontal cortex and its regulation by ethanol.
Bogenpohl JW, Weston RM, Foreman TN, Kitchen KE, Miles MF. Bogenpohl JW, et al. Alcohol Clin Exp Res. 2022 Jan;46(1):29-39. doi: 10.1111/acer.14754. Epub 2021 Dec 9. Alcohol Clin Exp Res. 2022. PMID: 34839533 Free PMC article. - Transcriptome analysis of chloride intracellular channel knockdown in Drosophila identifies oxidation-reduction function as possible mechanism of altered sensitivity to ethanol sedation.
Weston RM, Schmitt RE, Grotewiel M, Miles MF. Weston RM, et al. PLoS One. 2021 Jul 6;16(7):e0246224. doi: 10.1371/journal.pone.0246224. eCollection 2021. PLoS One. 2021. PMID: 34228751 Free PMC article. - Chloride intracellular channel proteins respond to heat stress in Caenorhabditis elegans.
Liang J, Shaulov Y, Savage-Dunn C, Boissinot S, Hoque T. Liang J, et al. PLoS One. 2017 Sep 8;12(9):e0184308. doi: 10.1371/journal.pone.0184308. eCollection 2017. PLoS One. 2017. PMID: 28886120 Free PMC article. - Drosophila and Caenorhabditis elegans as Discovery Platforms for Genes Involved in Human Alcohol Use Disorder.
Grotewiel M, Bettinger JC. Grotewiel M, et al. Alcohol Clin Exp Res. 2015 Aug;39(8):1292-311. doi: 10.1111/acer.12785. Epub 2015 Jul 14. Alcohol Clin Exp Res. 2015. PMID: 26173477 Free PMC article. Review. - The genetics of behavioral alcohol responses in Drosophila.
Rodan AR, Rothenfluh A. Rodan AR, et al. Int Rev Neurobiol. 2010;91:25-51. doi: 10.1016/S0074-7742(10)91002-7. Int Rev Neurobiol. 2010. PMID: 20813239 Free PMC article. Review.
Cited by
- Case-only exome variation analysis of severe alcohol dependence using a multivariate hierarchical gene clustering approach.
Gentry AE, Alexander JC, Ahangari M, Peterson RE, Miles MF, Bettinger JC, Davies AG, Groteweil M, Bacanu SA, Kendler KS, Riley BP, Webb BT; VCU Alcohol Research Center working group. Gentry AE, et al. PLoS One. 2023 Apr 25;18(4):e0283985. doi: 10.1371/journal.pone.0283985. eCollection 2023. PLoS One. 2023. PMID: 37098020 Free PMC article. - Chloride intracellular channel 4 (CLIC4) expression profile in the mouse medial prefrontal cortex and its regulation by ethanol.
Bogenpohl JW, Weston RM, Foreman TN, Kitchen KE, Miles MF. Bogenpohl JW, et al. Alcohol Clin Exp Res. 2022 Jan;46(1):29-39. doi: 10.1111/acer.14754. Epub 2021 Dec 9. Alcohol Clin Exp Res. 2022. PMID: 34839533 Free PMC article. - Transcriptome analysis of chloride intracellular channel knockdown in Drosophila identifies oxidation-reduction function as possible mechanism of altered sensitivity to ethanol sedation.
Weston RM, Schmitt RE, Grotewiel M, Miles MF. Weston RM, et al. PLoS One. 2021 Jul 6;16(7):e0246224. doi: 10.1371/journal.pone.0246224. eCollection 2021. PLoS One. 2021. PMID: 34228751 Free PMC article. - Flying Together: Drosophila as a Tool to Understand the Genetics of Human Alcoholism.
Lathen DR, Merrill CB, Rothenfluh A. Lathen DR, et al. Int J Mol Sci. 2020 Sep 11;21(18):6649. doi: 10.3390/ijms21186649. Int J Mol Sci. 2020. PMID: 32932795 Free PMC article. Review. - Brain regional gene expression network analysis identifies unique interactions between chronic ethanol exposure and consumption.
Smith ML, Lopez MF, Wolen AR, Becker HC, Miles MF. Smith ML, et al. PLoS One. 2020 May 29;15(5):e0233319. doi: 10.1371/journal.pone.0233319. eCollection 2020. PLoS One. 2020. PMID: 32469986 Free PMC article.
References
- Ashley RH. Challenging accepted ion channel biology: p64 and the CLIC family of putative intracellular anion channel proteins (Review) Molecular membrane biology. 2003;20:1–11. - PubMed
- Berry KL, Bulow HE, Hall DH, Hobert O. A C. elegans CLIC-like protein required for intracellular tube formation and maintenance. Science (New York, N.Y. 2003;302:2134–2137. - PubMed
- Berry KL, Hobert O. Mapping functional domains of chloride intracellular channel (CLIC) proteins in vivo. Journal of molecular biology. 2006;359:1316–1333. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P20 AA017828/AA/NIAAA NIH HHS/United States
- U01 AA016667/AA/NIAAA NIH HHS/United States
- R01 AA020634/AA/NIAAA NIH HHS/United States
- F31 AA018615/AA/NIAAA NIH HHS/United States
- R01 AA016837/AA/NIAAA NIH HHS/United States
- U01 AA016662/AA/NIAAA NIH HHS/United States
- R01 AA016842/AA/NIAAA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases