RNase H and postreplication repair protect cells from ribonucleotides incorporated in DNA - PubMed (original) (raw)
RNase H and postreplication repair protect cells from ribonucleotides incorporated in DNA
Federico Lazzaro et al. Mol Cell. 2012.
Abstract
The chemical identity and integrity of the genome is challenged by the incorporation of ribonucleoside triphosphates (rNTPs) in place of deoxyribonucleoside triphosphates (dNTPs) during replication. Misincorporation is limited by the selectivity of DNA replicases. We show that accumulation of ribonucleoside monophosphates (rNMPs) in the genome causes replication stress and has toxic consequences, particularly in the absence of RNase H1 and RNase H2, which remove rNMPs. We demonstrate that postreplication repair (PRR) pathways-MMS2-dependent template switch and Pol ζ-dependent bypass-are crucial for tolerating the presence of rNMPs in the chromosomes; indeed, we show that Pol ζ efficiently replicates over 1-4 rNMPs. Moreover, cells lacking RNase H accumulate mono- and polyubiquitylated PCNA and have a constitutively activated PRR. Our findings describe a crucial function for RNase H1, RNase H2, template switch, and translesion DNA synthesis in overcoming rNTPs misincorporated during DNA replication, and may be relevant for the pathogenesis of Aicardi-Goutières syndrome.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
Graphical abstract
Figure 1
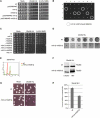
Abundant Incorporation of rNTPs into DNA Sensitizes Cells to Replication Stress and Is Lethal in Cells Lacking RNase H (A) To test sensitivity to sublethal doses of HU or MMS, 10-fold serial dilutions of the indicated mutant strains were plated on YPD, YPD + 25 mM HU and YPD + 0.04% MMS. Pictures were taken after 3 days of incubation. (B) Tetrads derived from a cross between rnh1Δ rnh201Δ and rnh1Δ pol2-M644G were dissected on YPD plates. Seven tetrads (1–7) are shown. The circles on the figure indicate the position of the original rhn1Δ rnh201Δ pol2-M644G spores. (C) Sensitivity to HU and MMS of the indicated strains was tested as described in (A). A checkpoint-defective mec1-1 strain was included as a positive control. (D) Single cells were isolated on YPD plates and grown for 22 hr in the presence of 25 mM HU; colonies were visualized by microscopic analysis. (E and F) wild-type and rnh1Δ rn201Δ cells were released in 25 mM HU after α factor arrest. After 180 min, cultures were analyzed by FACS, for DNA contents, and cell extracts were tested by western blotting with anti-Rad53 antibodies. (G) Wild-type and rnh1Δ rnh201Δ cells were plated on YPD with or without 25 mM HU in the presence of Phloxine B, which stains in red colonies containing dead cells. (H) Quantification of cell survival was obtained by plating G1 synchronized cells (100 cells per plate) on dishes containing 25 mM HU or mock. Colonies were counted after 3 days of incubation. The graph is representative of three independent experiments. Error bars describe standard deviation.
Figure 2
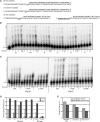
Postreplication Repair Is Specifically Required to Tolerate rNMPs-Containing Chromosomes Sensitivity to sublethal doses of HU was assayed as described in Figure 1. Pictures were taken after 3 days of incubation. The contribution of NER (A), BER (A), the two branches of PRR (B), and RAD51 (D) was tested. In (C), Quantification of cell survival was obtained as described in Figure 1H. The graph is representative of three independent experiments. Error bars describe standard deviation. It is worth noting that mms2Δ TLSΔ cells, despite being sensitive to HU in the spot tests, do not exhibit increased cell lethality, suggesting that the HU sensitivity derives from a very slow cell-cycle progression. In (E), tetrads derived from a cross between rnh1Δ rnh201Δ and rad52Δ were dissected on YPD plates. Five tetrads (1–5) are shown.The circles on the figure indicate the position of the original rhn1Δ rnh201Δ rad52Δ spores. Cells derived from such microcolonies do not grow when restreaked, revealing that a rad52Δ mutation is synthetic lethal with deletion of the RNH1 and RNH201 genes. TLSΔ comprises rev1Δ rev3Δ rev7Δ rad30Δ.
Figure 3
Pol ζ Allows Cells to Cope with Unrepaired rNMPs The sensitivity to HU was measured as described in Figure 1: the specific contribution of each TLS polymerase (A) and the requirement of the catalytic activity of Rev1 (B) were tested.
Figure 4
DNA Polymerase ζ Efficiently Bypasses rNMPs in the Template Strand (A) Primer-template sequences. In the 65-mer substrate, “x” is the position of the single rNMP (rG, rC, rA, or rU) and “g” is the position of the 5′-rG in the DNA template. In the 45-mer substrate, the underlined lowercase nucleotides indicate the position and sequence of the rNMPs in 4- and 16-rNMP substrates, respectively. (B and C) Phosphorimages of products generated during bypass of a single rNMP (B) and tracts of rNMPs by Pol δ and ζ (C). The template sequence is shown on the left, the arrow depicts the location of full-length product, and “r” represents the position of the rNMPs in the template. No enzyme was added to the unextended primer reaction (0 min). (D) Relative bypass efficiencies for Pols δ and ζ. Images of reaction products shown in (B) and (C) were quantified, and relative bypass efficiencies were calculated as described (Stone et al., 2009). The values for Pol δ with the 65-mer substrates in the absence of PCNA have been reported previously (Watt et al., 2011) and are shown here for comparison. The asterisks indicate the relative bypass values for Pol δ for reaction mixtures containing 200 nM PCNA. (E) Mutation rates for the pol2-M644G rnh201Δ and pol2-M644G rnh201Δ rev3Δ strains. The total mutation rates for resistance to 5-FOA were determined as described in Experimental Procedures. The 95% confidence intervals for the pol2-M644G rnh201Δ and pol2-M644G rnh201Δ rev3Δ strains were 110 to 200 and 57 to 140, respectively. For the pol2-M644G rnh201Δ strain, the rates for total 2–5 base pair deletions and for CA deletions at position 216–219 in URA3 are from Clark et al. (2011). For the pol2-M644G rnh201Δ rev3Δ strain, rates for short deletions were calculated after sequencing the ura3 gene in 163 independent 5-FOA resistant clones. Of these, 136 harbored 2–5 base pair deletions, 88 of which were CA deletions at the CACA hotspot at position 216–219 in URA3 (see spectrum in Figure S6).
Figure 5
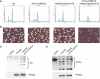
In the Absence of RNase H, the PRR Pathway Is Constitutively Activated and Promotes Cell Survival in an Unperturbed Cell Cycle The role of PRR was assessed in unperturbed rnh1Δ rnh201Δ cultures. Exponentially growing cells lacking RNase H and defective in PRR were analyzed by FACS (A), to monitor cell cycle distribution, and by Phloxine B staining (B), to evaluate cell lethality. PCNA was affinity purified from exponentially growing unperturbed wild-type cells or from cells lacking RNase H activity. PCNA levels were estimated by western blotting with anti-HIS Ab. PCNA ubiquitylation was monitored by western blotting with anti-ubiquitin Ab (C), and PCNA sumoylation was monitored by western blotting with anti-SUMO Ab (D).
Figure 6
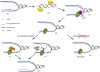
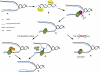
RNase H and Postreplication Repair Protect Cells from rNMPs Incorporated in Chromosomal DNA During DNA synthesis, replicative polymerases can incorporate rNTPs (red dot) in place of dNTPs (A). RNase H1 and RNase H2 are required to remove rNMPs from newly replicated DNA (blue line) (B). If rNMPs persist until the following cell cycle, they will create problems at during the DNA synthesis step (C), since replicative polymerases cannot efficiently elongate the nascent strand opposite rNMPs in the template strand (black line). Replication fork restart downstream of the lesions leaves incomplete replication products for postreplication repair (D). PCNA is ubiquitylated. Either MMS2-dependent template switch mechanisms (E) or Pol ζ-dependent translesion synthesis (F) allow bypass of rNMPs and completion of replication (G). Under these conditions, inactivation of PRR causes cell lethality (H).
Similar articles
- RNases H1 and H2: guardians of the stability of the nuclear genome when supply of dNTPs is limiting for DNA synthesis.
Cerritelli SM, El Hage A. Cerritelli SM, et al. Curr Genet. 2020 Dec;66(6):1073-1084. doi: 10.1007/s00294-020-01086-8. Epub 2020 Sep 4. Curr Genet. 2020. PMID: 32886170 Free PMC article. Review. - High density of unrepaired genomic ribonucleotides leads to Topoisomerase 1-mediated severe growth defects in absence of ribonucleotide reductase.
Cerritelli SM, Iranzo J, Sharma S, Chabes A, Crouch RJ, Tollervey D, El Hage A. Cerritelli SM, et al. Nucleic Acids Res. 2020 May 7;48(8):4274-4297. doi: 10.1093/nar/gkaa103. Nucleic Acids Res. 2020. PMID: 32187369 Free PMC article. - Reduction of hRNase H2 activity in Aicardi-Goutières syndrome cells leads to replication stress and genome instability.
Pizzi S, Sertic S, Orcesi S, Cereda C, Bianchi M, Jackson AP, Lazzaro F, Plevani P, Muzi-Falconi M. Pizzi S, et al. Hum Mol Genet. 2015 Feb 1;24(3):649-58. doi: 10.1093/hmg/ddu485. Epub 2014 Sep 30. Hum Mol Genet. 2015. PMID: 25274781 Free PMC article. - Elevated Genome-Wide Instability in Yeast Mutants Lacking RNase H Activity.
O'Connell K, Jinks-Robertson S, Petes TD. O'Connell K, et al. Genetics. 2015 Nov;201(3):963-75. doi: 10.1534/genetics.115.182725. Epub 2015 Sep 22. Genetics. 2015. PMID: 26400613 Free PMC article. - RNase H2-RED carpets the path to eukaryotic RNase H2 functions.
Cerritelli SM, Crouch RJ. Cerritelli SM, et al. DNA Repair (Amst). 2019 Dec;84:102736. doi: 10.1016/j.dnarep.2019.102736. Epub 2019 Oct 23. DNA Repair (Amst). 2019. PMID: 31761672 Free PMC article. Review.
Cited by
- Transcriptional responses to loss of RNase H2 in Saccharomyces cerevisiae.
Arana ME, Kerns RT, Wharey L, Gerrish KE, Bushel PR, Kunkel TA. Arana ME, et al. DNA Repair (Amst). 2012 Dec 1;11(12):933-41. doi: 10.1016/j.dnarep.2012.09.006. Epub 2012 Oct 15. DNA Repair (Amst). 2012. PMID: 23079308 Free PMC article. - Impact of Ribonucleotide Backbone on Translesion Synthesis and Repair of 7,8-Dihydro-8-oxoguanine.
Sassa A, Çağlayan M, Rodriguez Y, Beard WA, Wilson SH, Nohmi T, Honma M, Yasui M. Sassa A, et al. J Biol Chem. 2016 Nov 11;291(46):24314-24323. doi: 10.1074/jbc.M116.738732. Epub 2016 Sep 22. J Biol Chem. 2016. PMID: 27660390 Free PMC article. - A dual role of divalent metal ions in catalysis and folding of RNase H1 from extreme halophilic archaeon Halobacterium sp. NRC-1.
Tannous E, Yokoyama K, You DJ, Koga Y, Kanaya S. Tannous E, et al. FEBS Open Bio. 2012 Oct 27;2:345-52. doi: 10.1016/j.fob.2012.10.003. Print 2012. FEBS Open Bio. 2012. PMID: 23772368 Free PMC article. - Processing ribonucleotides incorporated during eukaryotic DNA replication.
Williams JS, Lujan SA, Kunkel TA. Williams JS, et al. Nat Rev Mol Cell Biol. 2016 Jun;17(6):350-63. doi: 10.1038/nrm.2016.37. Epub 2016 Apr 20. Nat Rev Mol Cell Biol. 2016. PMID: 27093943 Free PMC article. Review. - Determination of RNase H activity via real-time monitoring of target-triggered rolling circle amplification.
Lee CY, Kang KS, Park KS, Park HG. Lee CY, et al. Mikrochim Acta. 2017 Dec 14;185(1):53. doi: 10.1007/s00604-017-2610-8. Mikrochim Acta. 2017. PMID: 29594533
References
- Adams A., Gottschling D.E., Stearns T., Kaiser C.A. Cold Spring Harbor Laboratory Press; Plainview, NY: 1998. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual.
- Ayyagari R., Gomes X.V., Gordenin D.A., Burgers P.M. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 AND DNA2. J. Biol. Chem. 2003;278:1618–1625. - PubMed
- Ban C., Ramakrishnan B., Sundaralingam M. A single 2′-hydroxyl group converts B-DNA to A-DNA. Crystal structure of the DNA-RNA chimeric decamer duplex d(CCGGC)r(G)d(CCGG) with a novel intermolecular G-C base-paired quadruplet. J. Mol. Biol. 1994;236:275–285. - PubMed
- Burgers P.M., Gerik K.J. Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase delta. J. Biol. Chem. 1998;273:19756–19762. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GGP11003/TI_/Telethon/Italy
- NIHGM32431/PHS HHS/United States
- Z01 ES065070/Intramural NIH HHS/United States
- R01 GM032431/GM/NIGMS NIH HHS/United States
- 206281/ERC_/European Research Council/International
- CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous