Structural basis for recognition of H3T3ph and Smac/DIABLO N-terminal peptides by human Survivin - PubMed (original) (raw)
Structural basis for recognition of H3T3ph and Smac/DIABLO N-terminal peptides by human Survivin
Jiamu Du et al. Structure. 2012.
Abstract
Survivin is an inhibitor of apoptosis family protein implicated in apoptosis and mitosis. In apoptosis, it has been shown to recognize the Smac/DIABLO protein. It is also a component of the chromosomal passenger complex, a key player during mitosis. Recently, Survivin was identified in vitro and in vivo as the direct binding partner for phosphorylated Thr3 on histone H3 (H3T3ph). We have undertaken structural and binding studies to investigate the molecular basis underlying recognition of H3T3ph and Smac/DIABLO N-terminal peptides by Survivin. Our crystallographic studies establish recognition of N-terminal Ala in both complexes and identify intermolecular hydrogen-bonding interactions in the Survivin phosphate-binding pocket that contribute to H3T3ph mark recognition. In addition, our calorimetric data establish that Survivin binds tighter to the H3T3ph-containing peptide relative to the N-terminal Smac/DIABLO peptide, and this preference can be reversed through structure-guided mutations that increase the hydrophobicity of the phosphate-binding pocket.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures
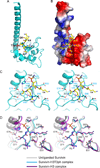
Figure 1. Interactions between H3(1–15)T3ph Peptide and Survivin in the Complex
- Overall interaction between residues Ala1 to Gln5 of H3(1–15)T3ph peptide and Survivin in the complex. The bound peptide (yellow) in a stick representation is positioned within the BIR domain (cyan) of Survivin in a ribbon representation.
- An electrostatics surface representation of Survivin with bound residues Ala1 to Gln5 of H3(1–15)T3ph peptide (yellow) in a stick representation. The N-terminus and Ala1 insert into a negatively charged pocket, the Arg2 and Lys4 side chains lie on a flat negatively charge surface, while Thr3ph lies on a positively charged shallow cleft. There is electrostatic complementarity between the bound H3T3ph peptide and Survivin in the complex.
- Stereo view highlighting details of the intermolecular interactions between the A1-R2-T3ph-K4 segment of the bound H3(1–15)T3ph peptide and binding pocket residues of the BIR domain of Survivin. Intermolecular hydrogen-bonding interactions are designated by dashed red lines.
- Stereo view of superpositioned structures of unliganded Survivin (grey), H3(1–15)T3ph peptide-bound Survivin (cyan) and H3(1–10) peptide-bound Survivin (magenta). These views emphasize binding pocket interactions. Note the shift in the α-helix (on left) on complex formation. See also Supplementary Figures S1, S2, S4, S5, S6 and S7.
Figure 2. Interactions between SmacN(1–15) Peptide and Survivin in the Complex
- Overall interaction between residues Ala1 to Ile4 of SmacN(1–15) peptide and Survivin in the complex. The bound peptide (yellow) in a stick representation is positioned within the BIR domain (cyan) of Survivin in a ribbon representation.
- An electrostatics surface representation of Survivin with bound residues Ala1 to Ile4 of SmacN peptide (yellow) in a stick representation.
- Stereo view highlighting details of the intermolecular interactions between the A1-V2-P3-I4 segment of the bound SmacN(1–15) peptide and binding pocket residues of the BIR domain of Survivin. Intermolecular hydrogen-bonding interactions are designated by dashed red lines.
- Stereo view of superpositioned structures of unliganded Survivin (grey) and SmacN peptide-bound Survivin (cyan). These views emphasize binding pocket interactions. Note the shift in the α-helix (on left) on complex formation.
See also Supplementary Figures S2 and S7.
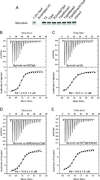
Figure 3. Pull-down and Isothermal Titration Calorimetry (ITC) Measurements of Binding Affinities between Survivin and H3 Peptides containing Modifications
- A pull-down assay testing binding of human Survivin to histone H3 containing different modifications. Purified human Survivin (1–142) was incubated with the indicated peptide beads. Coomassie staining of input and bead fractions is shown.
- ITC binding curve for complex formation between H3(1–15)T3ph peptide and Survivin.
- ITC binding curve for complex formation between H3(1–10) peptide and Survivin.
- ITC binding curve for complex formation between H3(1–20)R2me2aT3ph peptide and Survivin.
- ITC binding curve for complex formation between H3(1–20)T3phK4me3 peptide and Survivin.
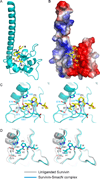
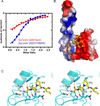
Figure 4. Interactions between SmacN(1–15) Peptide and Survivin (K62Y/H80W) Mutants in the Complex
- ITC binding curves for complex formation between SmacN(1–15) peptide and wild-type Survivin (red curve) and Survivin (K62Y/H80W) mutant (blue curve).
- An electrostatics surface representation of Survivin (K62R/H80W) mutant with bound residues A1 to I4 of SmacN(1–15) peptide (yellow) in a stick representation. The N-terminus and Ala1 insert into a negatively charged pocket, while the Pro3 ring lies on a hydrophobic surface formed by Tyr62 and Trp80.
- Stereo view highlighting details of the intermolecular interactions between the A1-V2-P3-I4 segment of the bound SmacN(1–15) peptide and binding pocket residues of the BIR domain of Survivin (K62R/H80W) mutant. Intermolecular hydrogen-bonding interactions are designated by dashed red lines.
See also Supplementary Figure 3.
Similar articles
- Interaction of SMAC with a survivin-derived peptide alters essential cancer hallmarks: Tumor growth, inflammation, and immunosuppression.
Santhanam M, Kumar Pandey S, Shteinfer-Kuzmine A, Paul A, Abusiam N, Zalk R, Shoshan-Barmatz V. Santhanam M, et al. Mol Ther. 2024 Jun 5;32(6):1934-1955. doi: 10.1016/j.ymthe.2024.04.007. Epub 2024 Apr 5. Mol Ther. 2024. PMID: 38582961 - Structural basis for the recognition of phosphorylated histone h3 by the survivin subunit of the chromosomal passenger complex.
Jeyaprakash AA, Basquin C, Jayachandran U, Conti E. Jeyaprakash AA, et al. Structure. 2011 Nov 9;19(11):1625-34. doi: 10.1016/j.str.2011.09.002. Epub 2011 Oct 25. Structure. 2011. PMID: 22032967 - Protein-protein recognition as a first step towards the inhibition of XIAP and Survivin anti-apoptotic proteins.
Obiol-Pardo C, Granadino-Roldán JM, Rubio-Martinez J. Obiol-Pardo C, et al. J Mol Recognit. 2008 May-Jun;21(3):190-204. doi: 10.1002/jmr.887. J Mol Recognit. 2008. PMID: 18438971 - Role of Smac/DIABLO in cancer progression.
Martinez-Ruiz G, Maldonado V, Ceballos-Cancino G, Grajeda JP, Melendez-Zajgla J. Martinez-Ruiz G, et al. J Exp Clin Cancer Res. 2008 Sep 26;27(1):48. doi: 10.1186/1756-9966-27-48. J Exp Clin Cancer Res. 2008. PMID: 18822137 Free PMC article. Review. - [Role of mitochondrial protein Smac/Diablo in regulation of apoptotic pathways].
Korga A, Korobowicz E, Dudka J. Korga A, et al. Pol Merkur Lekarski. 2006 May;20(119):573-6. Pol Merkur Lekarski. 2006. PMID: 16875166 Review. Polish.
Cited by
- Protocol for validating liquid-liquid phase separation as a driver of membraneless organelle assembly in vitro and in human cells.
Hedtfeld M, Musacchio A. Hedtfeld M, et al. STAR Protoc. 2024 Oct 23;5(4):103410. doi: 10.1016/j.xpro.2024.103410. Online ahead of print. STAR Protoc. 2024. PMID: 39446580 Free PMC article. - Molecular Insights into the Anticancer Activity of Withaferin-A: The Inhibition of Survivin Signaling.
Wadhwa R, Wang J, Shefrin S, Zhang H, Sundar D, Kaul SC. Wadhwa R, et al. Cancers (Basel). 2024 Sep 5;16(17):3090. doi: 10.3390/cancers16173090. Cancers (Basel). 2024. PMID: 39272948 Free PMC article. - Survivin as a Therapeutic Target for the Treatment of Human Cancer.
Wang Q, Greene MI. Wang Q, et al. Cancers (Basel). 2024 Apr 27;16(9):1705. doi: 10.3390/cancers16091705. Cancers (Basel). 2024. PMID: 38730657 Free PMC article. Review. - Histone Readers and Their Roles in Cancer.
Wen H, Shi X. Wen H, et al. Cancer Treat Res. 2023;190:245-272. doi: 10.1007/978-3-031-45654-1_8. Cancer Treat Res. 2023. PMID: 38113004 Free PMC article. - Neferine Targets the Oncogenic Characteristics of Androgen-Dependent Prostate Cancer Cells via Inducing Reactive Oxygen Species.
Dasari S, Pathak N, Thomas A, Bitla S, Kumar R, Munirathinam G. Dasari S, et al. Int J Mol Sci. 2023 Sep 18;24(18):14242. doi: 10.3390/ijms241814242. Int J Mol Sci. 2023. PMID: 37762540 Free PMC article.
References
- Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat. Med. 1997;3:917–921. - PubMed
- Chantalat L, Skoufias DA, Kleman JP, Jung B, Dideberg O, Margolis RL. Crystal structure of human survivin reveals a bow tie-shaped dimer with two unusual alpha-helical extensions. Mol. Cell. 2000;6:183–189. - PubMed
- Elia AE, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, Mohammad D, Cantley LC, Smerdon SJ, Yaffe MB. The molecular basis for phospho-dependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 2003;115:83–95. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA008748/CA/NCI NIH HHS/United States
- P41 RR015301/RR/NCRR NIH HHS/United States
- R01 GM075249/GM/NIGMS NIH HHS/United States
- R01GM075249-06A1/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases