Immunotherapeutic potential of anti-human endogenous retrovirus-K envelope protein antibodies in targeting breast tumors - PubMed (original) (raw)
Comparative Study
. 2012 Feb 8;104(3):189-210.
doi: 10.1093/jnci/djr540. Epub 2012 Jan 12.
Kiera Rycaj, Joshua B Plummer, Ming Li, Bingnan Yin, Katherine Frerich, Jeremy G Garza, Jianjun Shen, Kevin Lin, Peisha Yan, Sharon A Glynn, Tiffany H Dorsey, Kelly K Hunt, Stefan Ambs, Gary L Johanning
Affiliations
- PMID: 22247020
- PMCID: PMC3274512
- DOI: 10.1093/jnci/djr540
Comparative Study
Immunotherapeutic potential of anti-human endogenous retrovirus-K envelope protein antibodies in targeting breast tumors
Feng Wang-Johanning et al. J Natl Cancer Inst. 2012.
Abstract
Background: The envelope (env) protein of the human endogenous retrovirus type K (HERV-K) family is commonly expressed on the surface of breast cancer cells. We assessed whether HERV-K env is a potential target for antibody-based immunotherapy of breast cancer.
Methods: We examined the expression of HERV-K env protein in various malignant (MDA-MB-231, MCF-7, SKBR3, MDA-MB-453, T47D, and ZR-75-1) and nonmalignant (MCF-10A and MCF-10AT) human breast cell lines by immunoblot, enzyme-linked immunosorbent assay, immunofluorescence staining, and flow cytometry. Anti-HERV-K env monoclonal antibodies (mAbs; 6H5, 4D1, 4E11, 6E11, and 4E6) were used to target expression of HERV-K, and antitumor effects were assessed by quantifying growth and apoptosis of breast cancer cells in vitro, and tumor growth in vivo in mice (n = 5 per group) bearing xenograft tumors. The mechanisms responsible for 6H5 mAb-mediated effects were investigated by microarray assays, flow cytometry, immunoblot, and immunofluorescence staining. The expression of HERV-K env protein was assessed in primary breast tumors (n = 223) by immunohistochemistry. All statistical tests were two-sided.
Results: The expression of HERV-K env protein in malignant breast cancer cell lines was substantially higher than nonmalignant breast cells. Anti-HERV-K-specific mAbs inhibited growth and induced apoptosis of breast cancer cells in vitro. Mice treated with 6H5 mAb showed statistically significantly reduced growth of xenograft tumors compared with mice treated with control immunoglobulin (control [mIgG] vs 6H5 mAb, for tumors originating from MDA-MB-231 cells, mean size = 1448.33 vs 475.44 mm(3); difference = 972.89 mm(3), 95% CI = 470.17 to 1475.61 mm(3); P < .001). Several proteins involved in the apoptotic signaling pathways were overexpressed in vitro in 6H5 mAb-treated malignant breast cells compared with mIgG-treated control. HERV-K expression was detected in 148 (66%) of 223 primary breast tumors, and a higher rate of lymph node metastasis was associated with HERV-K-positive compared with HERV-K-negative tumors (43% vs 23%, P = .003).
Conclusion: Monoclonal antibodies against HERV-K env protein show potential as novel immunotherapeutic agents for breast cancer therapy.
Figures
Figure 1
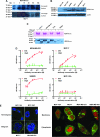
Analysis of envelope protein of human endogenous retrovirus type K (HERV-K env) expression in breast cancer cells. A)Metabolic turnover assay of biotin-pulsed HERV-K env in MDA-MB-231 and MCF-7 cells. The full-length and surface subunit env proteins of HERV-K were detected by immunoblot analysis using 6H5 monoclonal antibody (mAb). One representative blot of two independent experiments is shown. B) Immunoblot analysis of HERV-K env expression in breast cell lines. Protein lysates (50 μg per lane) from each cell line were loaded on each lane, and 6H5 mAb was used for detection. ACTB was used as the protein loading control. One representative blot of two independent experiments is shown. C) Deglycosylation analysis using peptide-N-glycosidase F (PNGase F). Cell lysates were treated with or without PNGase F. Glycosylated and deglycosylated HERV-K env proteins were detected using glycoprotein stained using periodic acid Schiff (top panel) or in an immunoblot analysis using 6H5 mAb (bottom panel). One representative blot of two independent experiments is shown. D) Surface expression of HERV-K env protein assessed on a panel of breast cell lines by cell enzyme-linked immunosorbent assay using 6H5 mAb as the primary antibody and mIgG as the negative control. OD = optical density. Results are representative of two independent experiments done. E) Immunofluorescent staining using 6H5 mAb showing the level of surface expression of HERV-K env protein (green fluorescence) on malignant (MCF-7 and MDA-MB-231) and nonmalignant (MCF-10A and MCF-10AT) breast cells (left panel). Scale bar = 20 μm. Surface (membrane) and cytoplasmic expression of HERV-K env protein (green fluorescence) in MCF-10A, MCF-7, MDA-MB-231, and MDA-MB-453 cells (right panel). Results are representative of two independent experiments. Scale bar = 20 μm. F) Flow cytometry assay of surface molecules of HERV-K env protein on breast cell lines. Cells were first treated with 6H5 mAb (red), followed by goat anti-mIgG-Alexa Fluor 647. Cells treated with isotoype control (IgG2a) followed by goat anti-mIgG-Alexa Fluor 647 treatment were used as control (gray). The mean fluorescence intensity of HERV-K for each cell line was calculated according to a calibration equation using quantitative indirect immunofluorescence beads (blue). Each of the five blue peaks from left to right represents a standard number of 1700, 11 000, 54 000, 194 000, and 561 000 beads, respectively. The mean fluorescence intensity of HERV-K was also compared between tumor cells (IDC51T) and matched uninvolved breast cells (IDC51N) obtained from a patient diagnosed with invasive ductal carcinoma (top panel). The number of surface molecules of HERV-K env protein in MCF-7, SKBR3, MDA-MB-231, and T47D cells treated with 6H5 mAb preincubated with K-GST (1 μg K-GST per 10 μg of 6H5 mAb) (blue) was compared with non-preincubated 6H5 mAb–treated cells (red) (bottom panel). Results are representative of at least three independent flow cytometry assays.
Figure 1
Analysis of envelope protein of human endogenous retrovirus type K (HERV-K env) expression in breast cancer cells. A)Metabolic turnover assay of biotin-pulsed HERV-K env in MDA-MB-231 and MCF-7 cells. The full-length and surface subunit env proteins of HERV-K were detected by immunoblot analysis using 6H5 monoclonal antibody (mAb). One representative blot of two independent experiments is shown. B) Immunoblot analysis of HERV-K env expression in breast cell lines. Protein lysates (50 μg per lane) from each cell line were loaded on each lane, and 6H5 mAb was used for detection. ACTB was used as the protein loading control. One representative blot of two independent experiments is shown. C) Deglycosylation analysis using peptide-N-glycosidase F (PNGase F). Cell lysates were treated with or without PNGase F. Glycosylated and deglycosylated HERV-K env proteins were detected using glycoprotein stained using periodic acid Schiff (top panel) or in an immunoblot analysis using 6H5 mAb (bottom panel). One representative blot of two independent experiments is shown. D) Surface expression of HERV-K env protein assessed on a panel of breast cell lines by cell enzyme-linked immunosorbent assay using 6H5 mAb as the primary antibody and mIgG as the negative control. OD = optical density. Results are representative of two independent experiments done. E) Immunofluorescent staining using 6H5 mAb showing the level of surface expression of HERV-K env protein (green fluorescence) on malignant (MCF-7 and MDA-MB-231) and nonmalignant (MCF-10A and MCF-10AT) breast cells (left panel). Scale bar = 20 μm. Surface (membrane) and cytoplasmic expression of HERV-K env protein (green fluorescence) in MCF-10A, MCF-7, MDA-MB-231, and MDA-MB-453 cells (right panel). Results are representative of two independent experiments. Scale bar = 20 μm. F) Flow cytometry assay of surface molecules of HERV-K env protein on breast cell lines. Cells were first treated with 6H5 mAb (red), followed by goat anti-mIgG-Alexa Fluor 647. Cells treated with isotoype control (IgG2a) followed by goat anti-mIgG-Alexa Fluor 647 treatment were used as control (gray). The mean fluorescence intensity of HERV-K for each cell line was calculated according to a calibration equation using quantitative indirect immunofluorescence beads (blue). Each of the five blue peaks from left to right represents a standard number of 1700, 11 000, 54 000, 194 000, and 561 000 beads, respectively. The mean fluorescence intensity of HERV-K was also compared between tumor cells (IDC51T) and matched uninvolved breast cells (IDC51N) obtained from a patient diagnosed with invasive ductal carcinoma (top panel). The number of surface molecules of HERV-K env protein in MCF-7, SKBR3, MDA-MB-231, and T47D cells treated with 6H5 mAb preincubated with K-GST (1 μg K-GST per 10 μg of 6H5 mAb) (blue) was compared with non-preincubated 6H5 mAb–treated cells (red) (bottom panel). Results are representative of at least three independent flow cytometry assays.
Figure 2
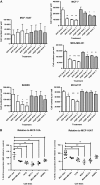
Effect of 6H5 monoclonal antibody (mAb) on growth of breast cells. A) Growth of MCF-10AT, MCF-7 MDA-MB-231, SKBR3, and IDCm73T breast cells after treatment with 10 μg/mL of 6H5 mAb. Cells were treated with 6H5 mAb or control mIgG for 24, 48, or 72 hours and counted after staining with trypan blue to determine the effect of antibody treatment on cell growth. Experiment was performed three times. Means and 95% confidence intervals (error bars) are presented. *P ≤ .05, **P ≤ .005, and ***P ≤ .001, compared with dose 0, calculated using two-sided Student's t test. B) DNA replication and cell cycle analysis by bromodeoxyuridine (BrdU) incorporation assay. Cells were treated with 10 μg/mL of either 6H5 mAb or mIgG (control) for 3 days and stained with 7-amino-actinomycin D coupled with immunofluorescent BrdU to determine the total cellular DNA in cells treated with mAbs. Each point represents a single measurement of BrdU incorporation. Experiment was performed three independent times. Means (horizontal bars) and 95% confidence intervals (error bars) are presented. *P ≤ .05, **P ≤ .005, and ***P ≤ .001, compared with MCF-10A or MCF-10AT, calculated using two-sided Student's t test.
Figure 3
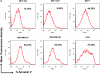
Effect of 6H5 monoclonal antibody (mAb) on apoptosis of breast cancer cells. A) Cells were treated with 6H5 mAb (red line) or control mIgG (gray line) (10 μg/mL of each antibody) for 16 hours, stained with annexin V–allophycocyanin and 7-AAD-phycoerythrin-cyanide 7, and analyzed by flow cytometry. B) Effect of 6H5 mAb treatment on cell death–inducing DFFA-like effector A (CIDEA) protein expression. Breast cancer cell lines were treated with 6H5 mAb or mIgG (10 μg/mL of each antibody) for 24 hours and analyzed for changes in protein expression by immunoblot using a mouse anti-human CIDEA antibody. ACTB was used as the protein loading control. Results are representative of two independent assays. C) The effect of 6H5 mAb treatment on expression of TP53 and TP53AIP1 proteins. MCF-7 and MDA-MB-231 breast cancer cell lines were treated with 6H5 mAb or mIgG (10 μg/mL of each antibody) for 24 hours, and an immunoblot assay was done using mouse anti-human TP53 and rabbit anti-human TP53AIP1 antibodies. ACTB was used as the protein loading control. Results are representative of two independent assays. D) Expression of active caspases 3 and 9 was assessed by immunoblot assay in ZR-75-1 and MDA-MB-231 breast cancer cells treated with 6H5 mAb or 6E11 mAb, or with mIgG (10 μg/mL of each antibody) for 24 hours using rabbit anti-human caspase 3 and mouse anti-human caspase 9 antibodies. ACTB was used as the protein loading control (top panel). Expression of active caspase 8 was assessed by immunoblot assay in MDA-MB-453 and MCF-7 breast cancer cells treated with 6H5 mAb (10 , 25, or 50 μg/mL), or with mIgG (10 μg/mL) for 24 hours using mouse anti-human caspase 8 antibody (bottom panel). Results are representative of at least two independent assays. E) Immunofluorescence assay to assess the expression of caspase proteins in MDA-MB-231 cells treated with 6H5 mAb or mIgG (10 μg/mL of each antibody) for 24 hours using rabbit anti-human caspase 3, mouse anti-human caspase 8, and mouse anti-human caspase 9 antibodies. Results are representative of two independent assays. Scale bar = 10 μm. F) Immunofluorescence assay to assess the expression of CDK5 and CDKN1A proteins. MDA-MB-231 cells were treated with 6H5 mAb or mIgG (10 μg/mL of each antibody) using mouse anti-human CDK5 and mouse anti-human CDKN1A antibodies. Results are representative of at least two independent assays. Scale bar = 10 μm.
Figure 3
Effect of 6H5 monoclonal antibody (mAb) on apoptosis of breast cancer cells. A) Cells were treated with 6H5 mAb (red line) or control mIgG (gray line) (10 μg/mL of each antibody) for 16 hours, stained with annexin V–allophycocyanin and 7-AAD-phycoerythrin-cyanide 7, and analyzed by flow cytometry. B) Effect of 6H5 mAb treatment on cell death–inducing DFFA-like effector A (CIDEA) protein expression. Breast cancer cell lines were treated with 6H5 mAb or mIgG (10 μg/mL of each antibody) for 24 hours and analyzed for changes in protein expression by immunoblot using a mouse anti-human CIDEA antibody. ACTB was used as the protein loading control. Results are representative of two independent assays. C) The effect of 6H5 mAb treatment on expression of TP53 and TP53AIP1 proteins. MCF-7 and MDA-MB-231 breast cancer cell lines were treated with 6H5 mAb or mIgG (10 μg/mL of each antibody) for 24 hours, and an immunoblot assay was done using mouse anti-human TP53 and rabbit anti-human TP53AIP1 antibodies. ACTB was used as the protein loading control. Results are representative of two independent assays. D) Expression of active caspases 3 and 9 was assessed by immunoblot assay in ZR-75-1 and MDA-MB-231 breast cancer cells treated with 6H5 mAb or 6E11 mAb, or with mIgG (10 μg/mL of each antibody) for 24 hours using rabbit anti-human caspase 3 and mouse anti-human caspase 9 antibodies. ACTB was used as the protein loading control (top panel). Expression of active caspase 8 was assessed by immunoblot assay in MDA-MB-453 and MCF-7 breast cancer cells treated with 6H5 mAb (10 , 25, or 50 μg/mL), or with mIgG (10 μg/mL) for 24 hours using mouse anti-human caspase 8 antibody (bottom panel). Results are representative of at least two independent assays. E) Immunofluorescence assay to assess the expression of caspase proteins in MDA-MB-231 cells treated with 6H5 mAb or mIgG (10 μg/mL of each antibody) for 24 hours using rabbit anti-human caspase 3, mouse anti-human caspase 8, and mouse anti-human caspase 9 antibodies. Results are representative of two independent assays. Scale bar = 10 μm. F) Immunofluorescence assay to assess the expression of CDK5 and CDKN1A proteins. MDA-MB-231 cells were treated with 6H5 mAb or mIgG (10 μg/mL of each antibody) using mouse anti-human CDK5 and mouse anti-human CDKN1A antibodies. Results are representative of at least two independent assays. Scale bar = 10 μm.
Figure 4
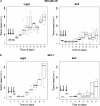
Antitumor effects of 6H5 monoclonal antibody (mAb) in immunodeficient mice carrying MDA-MB-231 xenograft tumors. A) Tumor sizes in MDA-MB-231 xenografts treated with 6H5 mAb (right panel) were compared with xenografts treated with mIgG (left panel) using a linear mixed-effects model. The solid arrows indicate the days of antibody injection. The tumor sizes of five mice on a particular day are shown, using a box and whiskers plot format. The horizontal line in the “box” indicates the median tumor size, the box represents interquartile range (25th and 75th percentiles), and the ends of the vertical lines or “whiskers” indicate the minimum and maximum data values. Open circles are outliers or suspected outliers. Error bars represent 95% confidence intervals from experiments performed two independent times with similar results. B) Tumor sizes were also assessed in MCF-7 xenografts treated with 6H5 mAb and compared with mIgG. Error bars represent with 95% confidence intervals from experiments performed two independent times with similar results. C) The number of TUNEL-positive (TUNEL+) cells in tumors from mice bearing MDA-MB-231 xenograft tumors treated with 6H5 mAb relative to mIgG-treated control tumors is shown. The horizontal bar represents the mean number of TUNEL+ cells in 6H5 mAb–treated and mIgG-treated tumors, and the solid circles represent positive cell counts in a single field (P = .008; Student's t test, two-sided) (left panel). Representative immunohistochemically stained sections of tumors from mIgG-treated and 6H5 mAb–treated mice are shown (right panel). Brown stain (TUNEL+) indicates apoptotic cells. These experiments were performed two independent times. Magnification = 400×. Scale bar = 200 μm. D) Ki-67-positive (Ki-67+) cell numbers were determined in xenograft tumors from mice injected with MDA-MB-231 cells and treated with 6H5 mAb compared with mIgG control. Representative immunohistochemical stains of tumors from 6H5 mAb–treated mice are shown: mIgG (right, top panel) and 6H5 mAb (right, bottom panel). The horizontal bar represents the mean number of Ki-67+cells in 6H5 mAb–treated and mIgG-treated tumors, and the solid circles represent positive cell counts in a single field (P < .001; calculated using a two-sided Student's t test) (left panel). These experiments were performed two independent times. Magnification = 400×. Scale bar = 200 μm.
Figure 4
Antitumor effects of 6H5 monoclonal antibody (mAb) in immunodeficient mice carrying MDA-MB-231 xenograft tumors. A) Tumor sizes in MDA-MB-231 xenografts treated with 6H5 mAb (right panel) were compared with xenografts treated with mIgG (left panel) using a linear mixed-effects model. The solid arrows indicate the days of antibody injection. The tumor sizes of five mice on a particular day are shown, using a box and whiskers plot format. The horizontal line in the “box” indicates the median tumor size, the box represents interquartile range (25th and 75th percentiles), and the ends of the vertical lines or “whiskers” indicate the minimum and maximum data values. Open circles are outliers or suspected outliers. Error bars represent 95% confidence intervals from experiments performed two independent times with similar results. B) Tumor sizes were also assessed in MCF-7 xenografts treated with 6H5 mAb and compared with mIgG. Error bars represent with 95% confidence intervals from experiments performed two independent times with similar results. C) The number of TUNEL-positive (TUNEL+) cells in tumors from mice bearing MDA-MB-231 xenograft tumors treated with 6H5 mAb relative to mIgG-treated control tumors is shown. The horizontal bar represents the mean number of TUNEL+ cells in 6H5 mAb–treated and mIgG-treated tumors, and the solid circles represent positive cell counts in a single field (P = .008; Student's t test, two-sided) (left panel). Representative immunohistochemically stained sections of tumors from mIgG-treated and 6H5 mAb–treated mice are shown (right panel). Brown stain (TUNEL+) indicates apoptotic cells. These experiments were performed two independent times. Magnification = 400×. Scale bar = 200 μm. D) Ki-67-positive (Ki-67+) cell numbers were determined in xenograft tumors from mice injected with MDA-MB-231 cells and treated with 6H5 mAb compared with mIgG control. Representative immunohistochemical stains of tumors from 6H5 mAb–treated mice are shown: mIgG (right, top panel) and 6H5 mAb (right, bottom panel). The horizontal bar represents the mean number of Ki-67+cells in 6H5 mAb–treated and mIgG-treated tumors, and the solid circles represent positive cell counts in a single field (P < .001; calculated using a two-sided Student's t test) (left panel). These experiments were performed two independent times. Magnification = 400×. Scale bar = 200 μm.
Figure 5
Expression of envelope protein of human endogenous retrovirus type K (HERV-K env) in invasive breast tumors from patients. Immunohistochemically stained sections were assessed in invasive breast tumors from four patients (representative of two independent experiments). HERV-K env was detected using 6H5 monoclonal antibody, shown as brown chromogen deposits. Top left tumor is negative for HERV-K env expression. Magnification = 100× for top right image and scale bar = 500 μm; magnification = 200× for all other images and scale bars = 200 μm.
Similar articles
- Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer.
Wang-Johanning F, Liu J, Rycaj K, Huang M, Tsai K, Rosen DG, Chen DT, Lu DW, Barnhart KF, Johanning GL. Wang-Johanning F, et al. Int J Cancer. 2007 Jan 1;120(1):81-90. doi: 10.1002/ijc.22256. Int J Cancer. 2007. PMID: 17013901 - CSPG4 protein as a new target for the antibody-based immunotherapy of triple-negative breast cancer.
Wang X, Osada T, Wang Y, Yu L, Sakakura K, Katayama A, McCarthy JB, Brufsky A, Chivukula M, Khoury T, Hsu DS, Barry WT, Lyerly HK, Clay TM, Ferrone S. Wang X, et al. J Natl Cancer Inst. 2010 Oct 6;102(19):1496-512. doi: 10.1093/jnci/djq343. Epub 2010 Sep 17. J Natl Cancer Inst. 2010. PMID: 20852124 Free PMC article. - Activation of HERV-K Env protein is essential for tumorigenesis and metastasis of breast cancer cells.
Zhou F, Li M, Wei Y, Lin K, Lu Y, Shen J, Johanning GL, Wang-Johanning F. Zhou F, et al. Oncotarget. 2016 Dec 20;7(51):84093-84117. doi: 10.18632/oncotarget.11455. Oncotarget. 2016. PMID: 27557521 Free PMC article. - Molecular biology of type A endogenous retrovirus.
Ono M. Ono M. Kitasato Arch Exp Med. 1990 Sep;63(2-3):77-90. Kitasato Arch Exp Med. 1990. PMID: 1710682 Review. - A new therapeutic approach for type 1 diabetes: Rationale for GNbAC1, an anti-HERV-W-Env monoclonal antibody.
Curtin F, Bernard C, Levet S, Perron H, Porchet H, Médina J, Malpass S, Lloyd D, Simpson R; RAINBOW-T1D investigators. Curtin F, et al. Diabetes Obes Metab. 2018 Sep;20(9):2075-2084. doi: 10.1111/dom.13357. Epub 2018 Jun 10. Diabetes Obes Metab. 2018. PMID: 29749030 Review.
Cited by
- Evolution of Repetitive Elements, Their Roles in Homeostasis and Human Disease, and Potential Therapeutic Applications.
Snowbarger J, Koganti P, Spruck C. Snowbarger J, et al. Biomolecules. 2024 Oct 2;14(10):1250. doi: 10.3390/biom14101250. Biomolecules. 2024. PMID: 39456183 Free PMC article. Review. - Expression of most retrotransposons in human blood correlates with biological aging.
Tsai YT, Seymen N, Thompson IR, Zou X, Mumtaz W, Gerlevik S, Mufti GJ, Karimi MM. Tsai YT, et al. Elife. 2024 Oct 17;13:RP96575. doi: 10.7554/eLife.96575. Elife. 2024. PMID: 39417397 Free PMC article. - Revolutionizing cancer treatment: an in-depth exploration of CAR-T cell therapies.
Kandav G, Chandel A. Kandav G, et al. Med Oncol. 2024 Oct 14;41(11):275. doi: 10.1007/s12032-024-02491-6. Med Oncol. 2024. PMID: 39400611 Review. - Prospects for breast cancer immunotherapy using microRNAs and transposable elements as objects.
Mustafin RN. Mustafin RN. Explor Target Antitumor Ther. 2024;5(5):1011-1026. doi: 10.37349/etat.2024.00261. Epub 2024 Aug 6. Explor Target Antitumor Ther. 2024. PMID: 39351441 Free PMC article. Review. - Regression of renal cell carcinoma by T cell receptor-engineered T cells targeting a human endogenous retrovirus.
Barisic S, Brahmbhatt EM, Cherkasova E, Spear TT, Savani U, Pierre S, Scurti GM, Chen L, Igboko M, Nadal R, Zeng G, Parry G, Stroncek DF, Highfill S, Dalheim AV, Reger R, Nishimura MI, Childs RW. Barisic S, et al. J Immunother Cancer. 2024 Sep 11;12(9):e009147. doi: 10.1136/jitc-2024-009147. J Immunother Cancer. 2024. PMID: 39266213 Free PMC article.
References
- Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. - PubMed
- Cho K, Lee YK, Greenhalgh DG. Endogenous retroviruses in systemic response to stress signals. Shock. Aug;30(2):105–116. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous