Both common variations and rare non-synonymous substitutions and small insertion/deletions in CLU are associated with increased Alzheimer risk - PubMed (original) (raw)
doi: 10.1186/1750-1326-7-3.
Nathalie Brouwers, Sebastiaan Engelborghs, Jean-Charles Lambert, Ekaterina Rogaeva, Rik Vandenberghe, Nathalie Le Bastard, Florence Pasquier, Steven Vermeulen, Jasper Van Dongen, Maria Mattheijssens, Karin Peeters, Richard Mayeux, Peter St George-Hyslop, Philippe Amouyel, Peter P De Deyn, Kristel Sleegers, Christine Van Broeckhoven
Affiliations
- PMID: 22248099
- PMCID: PMC3296573
- DOI: 10.1186/1750-1326-7-3
Both common variations and rare non-synonymous substitutions and small insertion/deletions in CLU are associated with increased Alzheimer risk
Karolien Bettens et al. Mol Neurodegener. 2012.
Abstract
Background: We have followed-up on the recent genome-wide association (GWA) of the clusterin gene (CLU) with increased risk for Alzheimer disease (AD), by performing an unbiased resequencing of all CLU coding exons and regulatory regions in an extended Flanders-Belgian cohort of Caucasian AD patients and control individuals (n = 1930). Moreover, we have replicated genetic findings by targeted resequencing in independent Caucasian cohorts of French (n = 2182) and Canadian (n = 573) origin and by performing meta-analysis combining our data with previous genetic CLU screenings.
Results: In the Flanders-Belgian cohort, we identified significant clustering in exons 5-8 of rare genetic variations leading to non-synonymous substitutions and a 9-bp insertion/deletion affecting the CLU β-chain (p = 0.02). Replicating this observation by targeted resequencing of CLU exons 5-8 in 2 independent Caucasian cohorts of French and Canadian origin identified identical as well as novel non-synonymous substitutions and small insertion/deletions. A meta-analysis, combining the datasets of the 3 cohorts with published CLU sequencing data, confirmed that rare coding variations in the CLU β-chain were significantly enriched in AD patients (OR(MH) = 1.96 [95% CI = 1.18-3.25]; p = 0.009). Single nucleotide polymorphisms (SNPs) association analysis indicated the common AD risk association (GWA SNP rs11136000, p = 0.013) in the 3 combined datasets could not be explained by the presence of the rare coding variations we identified. Further, high-density SNP mapping in the CLU locus mapped the common association signal to a more 5' CLU region.
Conclusions: We identified a new genetic risk association of AD with rare coding CLU variations that is independent of the 5' common association signal identified in the GWA studies. At this stage the role of these coding variations and their likely effect on the β-chain domain and CLU protein functioning remains unclear and requires further studies.
Figures
Figure 1
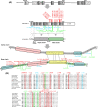
Schematic location of rare CLU coding variants identified in stage I, II and III resequencing. (A) Schematic presentation of CLU gene structure, CLU transcript 1 [NM_001831.2] and CLU protein [NP_001822.2]. Coding variants observed in AD patients only are indicated in red, variants observed in patients and controls in blue, variants detected in control individuals only in green. All predicted pathogenic variants are indicated in bold. After cleavage of the signal peptide, the secreted CLU form (449 AA) contains two coiled-coiled domains (pink), three amphipathic domains (blue) and a cysteine rich region (yellow) with 5 disulfide bridges (grey). Six N-glycosylation sites are marked in purple. For ease of interpretation, amino acids are given for specific CLU domains and for detected protein variants only. (B) Conservation alignment of amino acids of CLU beta-chain variants is shown in different species; Homo sapiens (ENSP00000315130), Gorilla gorilla (ENSGGOP00000016521), Pan troglodytes (ENSPTRP00000034423), Pongo abelii (ENSPPYP00000020696), Macaca mulata (ENSMMUP0000003216), Nomascus leucogenys (ENSNLEP00000020015), Tarsius syrichta (ENSTSYP00000001230), Mus musculus (ENSMUSP00000022616), Rattus norvegicus (ENSRNOP00000022095), Canis lupus familiaris (ENSCAFP00000012350) and Bos taurus (ENSBTAP00000007324). Similar to panel A, patient specific variants are marked in red, variants observed in patients and controls in blue and variants in control individuals in green. All predicted pathogenic variants are marked in bold.
Similar articles
- Association between CLU gene rs11136000 polymorphism and Alzheimer's disease: an updated meta-analysis.
Zhu R, Liu X, He Z. Zhu R, et al. Neurol Sci. 2018 Apr;39(4):679-689. doi: 10.1007/s10072-018-3259-8. Epub 2018 Feb 2. Neurol Sci. 2018. PMID: 29396813 - The CLU gene rs11136000 variant is significantly associated with Alzheimer's disease in Caucasian and Asian populations.
Liu G, Wang H, Liu J, Li J, Li H, Ma G, Jiang Y, Chen Z, Zhao B, Li K. Liu G, et al. Neuromolecular Med. 2014 Mar;16(1):52-60. doi: 10.1007/s12017-013-8250-1. Epub 2013 Jul 28. Neuromolecular Med. 2014. PMID: 23892938 - Reduced secreted clusterin as a mechanism for Alzheimer-associated CLU mutations.
Bettens K, Vermeulen S, Van Cauwenberghe C, Heeman B, Asselbergh B, Robberecht C, Engelborghs S, Vandenbulcke M, Vandenberghe R, De Deyn PP, Cruts M, Van Broeckhoven C, Sleegers K. Bettens K, et al. Mol Neurodegener. 2015 Jul 16;10:30. doi: 10.1186/s13024-015-0024-9. Mol Neurodegener. 2015. PMID: 26179372 Free PMC article. - Association of clusterin (CLU) variants and exfoliation syndrome: An analysis in two Caucasian studies and a meta-analysis.
Fan BJ, Pasquale LR, Kang JH, Levkovitch-Verbin H, Haines JL, Wiggs JL. Fan BJ, et al. Exp Eye Res. 2015 Oct;139:115-22. doi: 10.1016/j.exer.2015.08.004. Epub 2015 Aug 10. Exp Eye Res. 2015. PMID: 26272660 Free PMC article. Review. - Correlation of rs9331888 polymorphism with Alzheimer's disease among Caucasian and Chinese populations: a meta-analysis and systematic review.
Zhu B, Wang RM, Wang JT, Chen RL, Zheng YF, Zhang L, Zhao ZG. Zhu B, et al. Metab Brain Dis. 2017 Aug;32(4):981-989. doi: 10.1007/s11011-017-9957-8. Epub 2017 Feb 6. Metab Brain Dis. 2017. PMID: 28168383 Review.
Cited by
- Analyzing 74,248 Samples Confirms the Association Between CLU rs11136000 Polymorphism and Alzheimer's Disease in Caucasian But Not Chinese population.
Han Z, Qu J, Zhao J, Zou X. Han Z, et al. Sci Rep. 2018 Jul 23;8(1):11062. doi: 10.1038/s41598-018-29450-2. Sci Rep. 2018. PMID: 30038359 Free PMC article. - The Reelin Receptors Apolipoprotein E receptor 2 (ApoER2) and VLDL Receptor.
Dlugosz P, Nimpf J. Dlugosz P, et al. Int J Mol Sci. 2018 Oct 9;19(10):3090. doi: 10.3390/ijms19103090. Int J Mol Sci. 2018. PMID: 30304853 Free PMC article. Review. - Combined effects of Alzheimer risk variants in the CLU and ApoE genes on ventricular expansion patterns in the elderly.
Roussotte FF, Gutman BA, Madsen SK, Colby JB, Thompson PM; Alzheimer's Disease Neuroimaging Initiative. Roussotte FF, et al. J Neurosci. 2014 May 7;34(19):6537-45. doi: 10.1523/JNEUROSCI.5236-13.2014. J Neurosci. 2014. PMID: 24806679 Free PMC article. - Phenotypic characteristics of Alzheimer patients carrying an ABCA7 mutation.
Van den Bossche T, Sleegers K, Cuyvers E, Engelborghs S, Sieben A, De Roeck A, Van Cauwenberghe C, Vermeulen S, Van den Broeck M, Laureys A, Peeters K, Mattheijssens M, Vandenbulcke M, Vandenberghe R, Martin JJ, De Deyn PP, Cras P, Van Broeckhoven C; Belgian Neurology Consortium. Van den Bossche T, et al. Neurology. 2016 Jun 7;86(23):2126-33. doi: 10.1212/WNL.0000000000002628. Epub 2016 Apr 1. Neurology. 2016. PMID: 27037232 Free PMC article. - Clusterin in Alzheimer's Disease: Mechanisms, Genetics, and Lessons From Other Pathologies.
Foster EM, Dangla-Valls A, Lovestone S, Ribe EM, Buckley NJ. Foster EM, et al. Front Neurosci. 2019 Feb 28;13:164. doi: 10.3389/fnins.2019.00164. eCollection 2019. Front Neurosci. 2019. PMID: 30872998 Free PMC article. Review.
References
- Online Mendelian Inheritance in Man (OMIMI) database. http://www.ncbi.nlm.nih.gov/Omim
Publication types
MeSH terms
Substances
Grants and funding
- CAPMC/ CIHR/Canada
- HHMI/Howard Hughes Medical Institute/United States
- 089703/WT_/Wellcome Trust/United Kingdom
- MC_G1000734/MRC_/Medical Research Council/United Kingdom
- 081864/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous