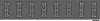Unusual N-terminal ααβαββα fold of PilQ from Thermus thermophilus mediates ring formation and is essential for piliation - PubMed (original) (raw)
Unusual N-terminal ααβαββα fold of PilQ from Thermus thermophilus mediates ring formation and is essential for piliation
Janin Burkhardt et al. J Biol Chem. 2012.
Abstract
DNA translocators of natural transformation systems are complex systems critical for the uptake of free DNA and provide a powerful mechanism for adaptation to changing environmental conditions. In natural transformation machineries, outer membrane secretins are suggested to form a multimeric pore for the uptake of external DNA. Recently, we reported on a novel structure of the DNA translocator secretin complex, PilQ, in Thermus thermophilus HB27 comprising a stable cone and cup structure and six ring structures with a large central channel. Here, we report on structural and functional analyses of a set of N-terminal PilQ deletion derivatives in T. thermophilus HB27. We identified 136 N-terminal residues exhibiting an unusual ααβαββα fold as a ring-building domain. Deletion of this domain had a dramatic effect on twitching motility, adhesion, and piliation but did not abolish natural transformation. These findings provide clear evidence that the pilus structures of T. thermophilus are not essential for natural transformation. The truncated complex was not affected in inner and outer membrane association, indicating that the 136 N-terminal residues are not essential for membrane targeting. Analyses of complex formation of the truncated PilQ monomers revealed that the region downstream of residue 136 is required for multimerization, and the region downstream of residue 207 is essential for monomer stability. Possible implications of our findings for the mechanism of DNA uptake are discussed.
Figures
FIGURE 1.
In trans complementation of pilQ deletion mutant HB27 Δ_pilQ_::bleo. A, the pilQ gene was amplified by PCR and cloned into pDM12 using restriction sites NdeI and NotI. The resulting plasmid pDM12-pilQhis-Q was electroporated into HB27 Δ_pilQ_::bleo. B, crude extracts of HB27 wild type (lane WT), HB27 Δ_pilQ_::bleo (lane Δ_Q_), and HB27 Δ_pilQ_::bleo carrying pDM12-pilQhis-Q (lane Q) were subjected to 3–12% polyacrylamide gradient SDS-PAGE followed by Western blot analysis. C, scheme of the putative secondary structure of PilQ and corresponding primer binding sites for the generation of N-terminal PilQ deletion derivatives by SDM using pDM12-pilQhis-Q as the template vector. The first maintained amino acid residues after SDM are indicated. Black boxes, α-helical domains; white boxes, β-sheets; aa, amino acid residues.
FIGURE 2.
Detection of N-terminal truncated and SDS-stable PilQ complexes. A, crude extracts of HB27 wild type (lane WT), HB27 Δ_pilQ_::bleo (lane Δ_Q_), and HB27 Δ_pilQ_::bleo carrying pDM12-pilQhis-Q (lane Q), -pilQΔ25–34his (lane 34), -pilQΔ25–64his (lane 64), -pilQΔ25–125his (lane 125), -pilQΔ25–207his (lane 207), or -pilQΔ25–262his (lane 262) were separated by 3–12% polyacrylamide gradient SDS-PAGE. PilQ production and SDS-stable complex assembly were verified by Western blot analysis. Arrows indicate expected PilQ derivative monomers. B, to identify potential SDS-unstable PilQ complexes, membranes of the indicated mutants were solubilized with 4% Triton-X-100, and solubilizates were subjected to 3–12% polyacrylamide gradient blue native PAGE followed by Western blot analysis.
FIGURE 3.
Identification of molecular mass of PilQ deletion derivatives by MALDI-MS. A, purified PilQ, PilQΔ25–64, and PilQΔ25–125 complexes were separated by blue native PAGE, electroeluted, and treated with hot phenol. These samples were subjected to SDS-PAGE and verified by immunoblotting using PilQ antibodies. B, purified PilQ, PilQΔ25–64, and PilQΔ25–125 complexes were dissociated using TFA and purified using ZipTips. The molecular mass of the monomers was analyzed by MALDI-MS. C, peptide mass fingerprinting was performed to characterize the sequence of PilQ, PilQΔ25–64, and PilQΔ25–125 monomers. Samples were prepared from SDS-polyacrylamide gels, digested using trypsin/chymotrypsin, and analyzed in a nano-HPLC-coupled electrospray ionization-quadrupole TOF mass spectrometer with data-dependent tandem MS spectrum acquisition. Matched peptides are shown in bold letters. Gray boxes, residues deleted by SDM; arrow, predicted signal peptide cleavage site. a.u., arbitrary units.
FIGURE 4.
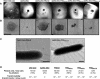
Electron microscopy analysis of PilQΔ25–64. Class averages after multivariate statistical analysis and classification of 800 particles reveal dimers of a truncated PilQ complex. Each class contains ∼65 images. Scale bar, 25 nm. 1–8, eight representative images of class averages.
FIGURE 5.
Colony morphology and cell adhesion on solid surfaces of T. thermophilus and HB27 Δ_pilQ_::bleo mutants. A, cells of T. thermophilus HB27 wild type, HB27 Δ_pilQ_::bleo, and HB27 Δ_pilQ_::bleo carrying pDM12-pilQhis-Q (PilQ-Q), -pilQΔ25–34his (PilQΔ25–34), -pilQΔ25–64his (PilQΔ25–64), or -pilQΔ25–125his (PilQΔ25–125) were stab-inoculated on minimal medium plates containing 1% BSA and incubated for 3 days at 68 °C under humid conditions. Colony morphology was documented under a binocular microscope. Scale bar, 5 mm. B, for analyzing cell adhesion and twitching motility, medium was removed from the Petri dish, and adhered cells were visualized by Coomassie staining. Scale bar, 10 mm. C, electron micrographs of piliated HB27 and non-piliated HB27 mutant cells. Scale bars, 0.5 μm. D, statistical analyses of piliation of the indicated complementation mutant cells by electron microscopy.
FIGURE 6.
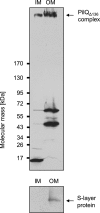
Subcellular localization of PilQΔ136 complex. Total membranes of HB27 Δ_pilQ_::bleo carrying pDM12-pilQΔ25–64his were separated into IM and outer membrane (OM) fractions. The fractions were subjected to SDS-PAGE in a 3–12% polyacrylamide gradient gel followed by immunoblotting using PilQ antibodies. The PilQ complex is indicated by an arrow. The purity of IM fractions was verified by Western blot analysis using S-layer protein-specific antibodies.
FIGURE 7.
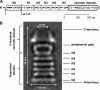
Secondary structure and subdomain prediction of T. thermophilus PilQ. The PredictProtein server was used to predict potentially α-helical (black boxes) and β-sheet (white boxes) regions within PilQ (A). The putative signal peptide processing site is indicated by an arrow, and the determined truncation site of the PilQΔ136 complex is displayed. Rounded boxes indicate putative N0, N1, N2, N3, N4, and N5 subdomains and the secretin domain. Subdomains are indicated in the PilQ complex structure (B). aa, amino acid(s).
Similar articles
- Cryo-EM structure of the bifunctional secretin complex of Thermus thermophilus.
D'Imprima E, Salzer R, Bhaskara RM, Sánchez R, Rose I, Kirchner L, Hummer G, Kühlbrandt W, Vonck J, Averhoff B. D'Imprima E, et al. Elife. 2017 Dec 27;6:e30483. doi: 10.7554/eLife.30483. Elife. 2017. PMID: 29280731 Free PMC article. - Structure and function of PilQ, a secretin of the DNA transporter from the thermophilic bacterium Thermus thermophilus HB27.
Burkhardt J, Vonck J, Averhoff B. Burkhardt J, et al. J Biol Chem. 2011 Mar 25;286(12):9977-84. doi: 10.1074/jbc.M110.212688. Epub 2011 Feb 1. J Biol Chem. 2011. PMID: 21285351 Free PMC article. - Topology and Structure/Function Correlation of Ring- and Gate-forming Domains in the Dynamic Secretin Complex of Thermus thermophilus.
Salzer R, D'Imprima E, Gold VA, Rose I, Drechsler M, Vonck J, Averhoff B. Salzer R, et al. J Biol Chem. 2016 Jul 8;291(28):14448-56. doi: 10.1074/jbc.M116.724153. Epub 2016 May 11. J Biol Chem. 2016. PMID: 27226590 Free PMC article. - Functional dissection of structural regions of the Thermus thermophilus competence protein PilW: Implication in secretin complex stability, natural transformation and pilus functions.
Yaman D, Averhoff B. Yaman D, et al. Biochim Biophys Acta Biomembr. 2021 Oct 1;1863(10):183666. doi: 10.1016/j.bbamem.2021.183666. Epub 2021 Jun 16. Biochim Biophys Acta Biomembr. 2021. PMID: 34143999 - Shuffling genes around in hot environments: the unique DNA transporter of Thermus thermophilus.
Averhoff B. Averhoff B. FEMS Microbiol Rev. 2009 May;33(3):611-26. doi: 10.1111/j.1574-6976.2008.00160.x. Epub 2009 Jan 16. FEMS Microbiol Rev. 2009. PMID: 19207744 Review.
Cited by
- Feature architecture aware phylogenetic profiling indicates a functional diversification of type IVa pili in the nosocomial pathogen Acinetobacter baumannii.
Iruegas R, Pfefferle K, Göttig S, Averhoff B, Ebersberger I. Iruegas R, et al. PLoS Genet. 2023 Jul 27;19(7):e1010646. doi: 10.1371/journal.pgen.1010646. eCollection 2023 Jul. PLoS Genet. 2023. PMID: 37498819 Free PMC article. - Cryo-EM structure of the bifunctional secretin complex of Thermus thermophilus.
D'Imprima E, Salzer R, Bhaskara RM, Sánchez R, Rose I, Kirchner L, Hummer G, Kühlbrandt W, Vonck J, Averhoff B. D'Imprima E, et al. Elife. 2017 Dec 27;6:e30483. doi: 10.7554/eLife.30483. Elife. 2017. PMID: 29280731 Free PMC article. - Type IV pilus biogenesis, twitching motility, and DNA uptake in Thermus thermophilus: discrete roles of antagonistic ATPases PilF, PilT1, and PilT2.
Salzer R, Joos F, Averhoff B. Salzer R, et al. Appl Environ Microbiol. 2014 Jan;80(2):644-52. doi: 10.1128/AEM.03218-13. Epub 2013 Nov 8. Appl Environ Microbiol. 2014. PMID: 24212586 Free PMC article. - Structure of a type IV pilus machinery in the open and closed state.
Gold VA, Salzer R, Averhoff B, Kühlbrandt W. Gold VA, et al. Elife. 2015 May 21;4:e07380. doi: 10.7554/eLife.07380. Elife. 2015. PMID: 25997099 Free PMC article. - Different effects of MglA and MglB on pilus-mediated functions and natural competence in Thermus thermophilus.
Salzer R, Joos F, Averhoff B. Salzer R, et al. Extremophiles. 2015 Mar;19(2):261-7. doi: 10.1007/s00792-014-0711-4. Epub 2014 Dec 4. Extremophiles. 2015. PMID: 25472010
References
- Lawrence J. G. (1999) Gene transfer, speciation, and the evolution of bacterial genomes. Curr. Opin. Microbiol. 2, 519–523 - PubMed
- Johnsborg O., Eldholm V., Håvarstein L. S. (2007) Natural genetic transformation: prevalence, mechanisms and function. Res. Microbiol. 158, 767–778 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources