Metaboloepigenetics: interrelationships between energy metabolism and epigenetic control of gene expression - PubMed (original) (raw)
Review
Metaboloepigenetics: interrelationships between energy metabolism and epigenetic control of gene expression
Dallas R Donohoe et al. J Cell Physiol. 2012 Sep.
Abstract
Diet and energy metabolism affect gene expression, which influences human health and disease. Here, we discuss the role of epigenetics as a mechanistic link between energy metabolism and control of gene expression. A number of key energy metabolites including SAM, acetyl-CoA, NAD(+), and ATP serve as essential co-factors for many, perhaps most, epigenetic enzymes that regulate DNA methylation, posttranslational histone modifications, and nucleosome position. The relative abundance of these energy metabolites allows a cell to sense its energetic state. And as co-factors, energy metabolites act as rheostats to modulate the activity of epigenetic enzymes and upregulate/downregulate transcription as appropriate to maintain homeostasis.
Copyright © 2012 Wiley Periodicals, Inc.
Figures
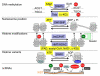
Figure 1. A simple model of metaboloepigenetics
The importance of food is not restricted to its nutritional content (e.g., calories and essential vitamins/minerals), but it is also important because bioactive food components and energy metabolites can function as environmental factors. And many gene-environment interactions converge at the level of the epigenome, which regulates gene expression profiles and determines phenotypic outcomes, referred to collectively as the phenome, at both the cellular and organismal levels. This is particularly true for certain energy metabolites and bioactive food components that activate or inhibit chromatinmodifying factors.
Figure 2. Epigenetic marks are catalyzed by chromatin-modifying factors
General types of epigenetic/epigenomic marks are listed at the left and a schematic of each reversible reaction is shown to the right. In the top 4 panels, enzymes are highlighted in blue and co-factors are highlighted in yellow. Nucleosome position and incorporation of histone variants are also reversible as indicated. In the bottom panel, XIST recruits enzymes and epigenomic marks in a sequential manner (from left to right) with stimulatory and inhibitory marks shown in green and red, respectively. See text for additional details.
Figure 3. 1C-cycle metabolism and SAM regulate DNA methylation
(A) Schematic of 1C cycle and synthesis of SAM. (B) Photograph of 5 genetically-identical agouti viable-yellow mice with different coat-color phenotypes ranging from yellow through 3 degrees of mottling to agouti. (C) Schematic of agouti-viable yellow “epialleles”. Agouti exons are shown as boxes with the filled portions corresponding to coding sequence. An IAP retrovirus-like element is immediately upstream of the agouti gene. In the top panel, CpG methylation (red circles labeled CH3) of the IAP renders it functionally inert. The endogenous promoter of the agouti gene (arrow) drives modest gene activity resulting in a brown (agouti) coat-color appearance in these mice. In the bottom panel, the IAP is not methylated and a strong, cryptic promoter (thicker arrow) drives high-level gene activity resulting in yellow pigmentation. (D) Methyl-donor supplementation provided to pregnant viable-yellow mice changes the coat-color distribution of their progeny after birth. Adapted from references 1 and .
Figure 4. TCA cycle intermediates serve as co-factors for epigenetic enzymes
Shown is a schematic of the TCA cycle. Several intermediates that are boxed or encircled (acetyl-Co-A, NAD+, α-ketoglutarate, and FAD) serve as essential co-factors for epigenetic enzymes (highlighted by colors).
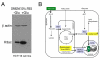
Figure 5. Glucose regulates histone acetylation
(A) Western blot showing pan-histone 3 acetylation (H3ac) levels in HCT116 cells grown in DMEM formulated without glucose (−Glu, left lane) or formulated with 25mM glucose (+Glu, right lane). β actin serves as the loading control. (B) A model of how glucose increases histone acetylation. First, it contributes carbons that can be incorporated into acetyl-CoA via the following pathway (denoted by thick arrows): glycolysis, PDH (pyruvate dehydrogenase), the TCA cycle and citrate shuttle, and ACL. Second, increased TCA-cycle activity increases the NADH/NAD+ ratio to inhibit SIRT1, which inhibits histone acetylation. Enzymes and metabolites that are most relevant are highlighted in blue and yellow, respectively.
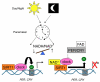
Figure 6. The redox state of NAD regulates SIRT1 and circadian oscillations
The day-night cycle sets an internal pacemaker that regulates circadian rhythms. Daily fluctuations in the NADH/NAD+ ratio contribute by regulating SIRT1 and its ability to counteract clock HAT activity. This helps determine histone acetylation and transcriptional activity of downstream target genes such as PER and CRY. See text for more details.
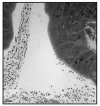
Figure 7. There is a preponderance of microbial cells in the lumen of the GI tract
Shown is a transmission electron micrograph of the proximal mouse colon. This image exemplifies the number and diversity of bacteria present in the mammalian GI tract. Bacterial metabolites such as butyrate can influence host epigeneitcs.
Similar articles
- Metabolism and epigenetics.
Janke R, Dodson AE, Rine J. Janke R, et al. Annu Rev Cell Dev Biol. 2015;31:473-496. doi: 10.1146/annurev-cellbio-100814-125544. Epub 2015 Sep 10. Annu Rev Cell Dev Biol. 2015. PMID: 26359776 Free PMC article. Review. - Crosstalk between metabolism and epigenetic modifications in autoimmune diseases: a comprehensive overview.
Wang Z, Long H, Chang C, Zhao M, Lu Q. Wang Z, et al. Cell Mol Life Sci. 2018 Sep;75(18):3353-3369. doi: 10.1007/s00018-018-2864-2. Epub 2018 Jul 4. Cell Mol Life Sci. 2018. PMID: 29974127 Free PMC article. Review. - Sink into the Epigenome: Histones as Repositories That Influence Cellular Metabolism.
Ye C, Tu BP. Ye C, et al. Trends Endocrinol Metab. 2018 Sep;29(9):626-637. doi: 10.1016/j.tem.2018.06.002. Epub 2018 Jul 11. Trends Endocrinol Metab. 2018. PMID: 30001904 Free PMC article. Review. - Methyl-Metabolite Depletion Elicits Adaptive Responses to Support Heterochromatin Stability and Epigenetic Persistence.
Haws SA, Yu D, Ye C, Wille CK, Nguyen LC, Krautkramer KA, Tomasiewicz JL, Yang SE, Miller BR, Liu WH, Igarashi K, Sridharan R, Tu BP, Cryns VL, Lamming DW, Denu JM. Haws SA, et al. Mol Cell. 2020 Apr 16;78(2):210-223.e8. doi: 10.1016/j.molcel.2020.03.004. Epub 2020 Mar 23. Mol Cell. 2020. PMID: 32208170 Free PMC article. - Metabolic regulation of histone post-translational modifications.
Fan J, Krautkramer KA, Feldman JL, Denu JM. Fan J, et al. ACS Chem Biol. 2015 Jan 16;10(1):95-108. doi: 10.1021/cb500846u. ACS Chem Biol. 2015. PMID: 25562692 Free PMC article. Review.
Cited by
- The role of neutrophils in trained immunity.
Kalafati L, Hatzioannou A, Hajishengallis G, Chavakis T. Kalafati L, et al. Immunol Rev. 2023 Mar;314(1):142-157. doi: 10.1111/imr.13142. Epub 2022 Oct 3. Immunol Rev. 2023. PMID: 36190144 Free PMC article. Review. - The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation.
Donohoe DR, Collins LB, Wali A, Bigler R, Sun W, Bultman SJ. Donohoe DR, et al. Mol Cell. 2012 Nov 30;48(4):612-26. doi: 10.1016/j.molcel.2012.08.033. Epub 2012 Oct 11. Mol Cell. 2012. PMID: 23063526 Free PMC article. - Memory Macrophages.
Kloc M, Kubiak JZ, Zdanowski R, Ghobrial RM. Kloc M, et al. Int J Mol Sci. 2022 Dec 20;24(1):38. doi: 10.3390/ijms24010038. Int J Mol Sci. 2022. PMID: 36613481 Free PMC article. Review. - Metabolic Signature of Energy Metabolism Alterations and Excess Nitric Oxide Production in Culture Media Correlate with Low Human Embryo Quality and Unsuccessful Pregnancy.
Pallisco R, Lazzarino G, Bilotta G, Marroni F, Mangione R, Saab MW, Brundo MV, Pittalà A, Caruso G, Capoccia E, Lazzarino G, Tavazzi B, Bilotta P, Amorini AM. Pallisco R, et al. Int J Mol Sci. 2023 Jan 3;24(1):890. doi: 10.3390/ijms24010890. Int J Mol Sci. 2023. PMID: 36614333 Free PMC article. - Immunometabolic Crosstalk: An Ancestral Principle of Trained Immunity?
Penkov S, Mitroulis I, Hajishengallis G, Chavakis T. Penkov S, et al. Trends Immunol. 2019 Jan;40(1):1-11. doi: 10.1016/j.it.2018.11.002. Epub 2018 Nov 29. Trends Immunol. 2019. PMID: 30503793 Free PMC article. Review.
References
- Prevention of neural tube defects: results of the Medical Research Council Vitamin Study. MRC Vitamin Study Research Group. Lancet. 1991;338(8760):131–137. - PubMed
- Burnett C, Valentini S, Cabreiro F, Goss M, Somogyvari M, Piper MD, Hoddinott M, Sutphin GL, Leko V, McElwee JJ, Vazquez-Manrique RP, Orfila AM, Ackerman D, Au C, Vinti G, Riesen M, Howard K, Neri C, Bedalov A, Kaeberlein M, Soti C, Partridge L, Gems D. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 477(7365):482–485. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources