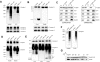YME1L controls the accumulation of respiratory chain subunits and is required for apoptotic resistance, cristae morphogenesis, and cell proliferation - PubMed (original) (raw)
YME1L controls the accumulation of respiratory chain subunits and is required for apoptotic resistance, cristae morphogenesis, and cell proliferation
Lukas Stiburek et al. Mol Biol Cell. 2012 Mar.
Abstract
Mitochondrial ATPases associated with diverse cellular activities (AAA) proteases are involved in the quality control and processing of inner-membrane proteins. Here we investigate the cellular activities of YME1L, the human orthologue of the Yme1 subunit of the yeast i-AAA complex, using stable short hairpin RNA knockdown and expression experiments. Human YME1L is shown to be an integral membrane protein that exposes its carboxy-terminus to the intermembrane space and exists in several complexes of 600-1100 kDa. The stable knockdown of YME1L in human embryonic kidney 293 cells led to impaired cell proliferation and apoptotic resistance, altered cristae morphology, diminished rotenone-sensitive respiration, and increased susceptibility to mitochondrial membrane protein carbonylation. Depletion of YME1L led to excessive accumulation of nonassembled respiratory chain subunits (Ndufb6, ND1, and Cox4) in the inner membrane. This was due to a lack of YME1L proteolytic activity, since the excessive accumulation of subunits was reversed by overexpression of wild-type YME1L but not a proteolytically inactive YME1L variant. Similarly, the expression of wild-type YME1L restored the lamellar cristae morphology of YME1L-deficient mitochondria. Our results demonstrate the importance of mitochondrial inner-membrane proteostasis to both mitochondrial and cellular function and integrity and reveal a novel role for YME1L in the proteolytic regulation of respiratory chain biogenesis.
Figures
FIGURE 1:
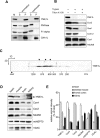
The characterization of human YME1L.(A) Human YME1L is an integral membrane protein. Submitochondrial fractions were prepared from HEK293 cell mitochondria (1 mg/ml) using either sonic disruption or extraction with 100 mM sodium carbonate (pH 11.5). The resulting supernatant (S) and pellet (P) fractions and untreated mitochondria (M) were immunoblotted with antibodies against YME1L, OXA1L, PNPase, and ATPase F1-α. (B) Human YME1L exposes its carboxy-terminus to the mitochondrial intermembrane space. Mitochondria isolated from HEK293 cells were swollen using hypotonic shock to selectively break the outer mitochondrial membrane and either left untreated or incubated with 1% Triton X-100 and/or 25 μg/ml trypsin for 20 min. After incubation, the trypsin activity was inhibited by soybean trypsin inhibitor, and mitochondria were immunoblotted with peptide-specific YME1L antiserum and with antibodies to Cox2, Sco1, Core 2, Cox1, and Ndufb6. (C) The determination of the native molecular weight of human YME1L using BN-PAGE. Mitochondria isolated from HEK293 cells were solubilized with 1% dodecyl maltoside, and ∼50 μg of protein was fractionated using two-dimensional blue native/denaturing PAGE on a 3–12% polyacrylamide gradient in the first dimension. The resulting immunoblots were incubated with antiserum raised against human YME1L. To enable the estimation of the native molecular weights of the observed YME1L complexes, the membrane was subsequently immunoblotted with antibodies to Ndufa9, ATPase F1-α, core 2, and Cox2. Under the conditions used, these antibodies recognize the complex I holoenzyme (970 kDa), ATP synthase holoenzyme (600 kDa), ATP synthase dimer (1200 kDa), F1-ATPase subcomplex (370 kDa), complex III dimer (500 kDa), and complex IV monomer (200 kDa), respectively. Black arrows denote the three major observed forms of the human i-AAA complex. (D) The expression level of human YME1L is tissue dependent. Mitochondria (25 μg of protein) isolated from control human cardiac muscle, skeletal muscle, frontal cortex, and kidney tissue were resolved using SDS–PAGE and immunoblotted with antibodies to YME1L, Cox4, Cox2, and Ndufb6. VDAC and mtHSP70 were used to control for actual mitochondrial enrichment in various preparations. The arrow denotes the migration of human YME1L isoform 3. (E) The densitometric quantification of immunoblot signals from D. The sum of signals from all four tissues for each protein was set to 100%. Error bars correspond to SD from the mean.
FIGURE 2:
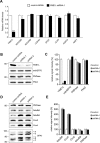
The loss of YME1L leads to the stabilization of the respiratory chain subunits Ndufb6 and Cox4 and to an altered pattern of OPA1 isoforms. (A) YME1L transcripts are efficiently depleted using stable shRNA knockdown. The relative quantification of the analyzed transcripts was performed with TaqMan Gene Expression Assays on a 7300 Real-Time PCR System (Applied Biosystems). HPRT1 (hypoxanthine phosphoribosyltransferase 1), TUBA1A (tubulin, alpha 1a), GAPDH (glyceraldehyde-3-phosphate dehydrogenase), and TBP (TATA box–binding protein) were used as reference genes. *p < 0.05, ***p < 0.001. (B) The loss of human YME1L protein does not affect mitochondrial PNPase or Phb2 levels. Equal amounts of mitochondrial lysates were separated (∼10 μg of protein) using SDS–PAGE and immunoblotted with antibodies to YME1L, mtHSP70, Phb2, and PNPase. (C) The densitometric quantification of the immunoblot signals in B. (D) Whereas Ndufb6, Cox4, and OPA1 are markedly affected by the loss of YME1L, the cellular levels of Cox2 and PNPase are unaffected. Equal amounts of whole-cell lysates (∼20 μg of protein) were separated using SDS–PAGE and immunoblotted with antibodies to PNPase, Ndufb8, Ndufb6, Cox2, Cox4, and OPA1. Controls correspond to HEK293 cells transfected with the scrambled shRNA.(E) The densitometric quantification of the immunoblot signals from (D), excluding the OPA1 signal. Error bars correspond to SD from the mean.
FIGURE 3:
Ndufb6, Cox4, and Cox2 are proteolytic substrates of human YME1L. (A) The expression of wild-type (wt) YME1L, but not the proteolytically inactive YME1LE543Q variant, reverses the stabilization of Ndufb6 and Cox4 in YME1L KD cells. YME1L KD cells were transiently transfected with the empty vector, the wild-type YME1L construct, or the YME1LE543Q-FLAG construct and, together with the scrambled shRNA-transformed HEK293 cells (control lane), used to prepare whole-cell lysates, which were subsequently immunoblotted with antibodies to Ndufb6, Cox4, Cox2, and SDHA. Signals were quantified by densitometric analysis. (B) Cox2 and Ndufb6 efficiently coimmunoprecipitate with proteolytically inactive YME1LE534Q-FLAG in YME1L KD cells. YME1L KD cells were transfected as in A and used to prepare mitochondrial fractions. Mitochondria were solubilized with 1% Triton X-100 and incubated with Anti-FLAG M2 affinity gel (Sigma-Aldrich). The eluted antigens, together with a fraction of vector-transfection input (5%), were immunoblotted with antibodies to Cox2, Cox5a, and Ndufb6 and with the monoclonal anti-FLAG M2 antibody. (C) YME1L isoform 3 is expressed in HEK293 cells. Endogenous YME1L was immunoprecipitated from HEK293 cell mitochondria (1 mg/ml) using anti-YME1L antiserum, resolved along with preimmune serum precipitate using SDS–PAGE, silver stained, and then excised and subjected to identification by mass fingerprinting and tandem mass spectrometry analysis. (D) Subunit d of complex V coimmunoprecipitates with the endogenous YME1L from HEK293 mitochondria, but Cox2 and Ndufb6 do not. The immunoprecipitate shown in C was immunoblotted with antibodies to ATP synthase subunit d, Cox2, and Ndufb6. (E) The densitometric quantification of immunoblot signals from A. Error bars correspond to SD from the mean.
FIGURE 4:
Excess Ndufb6 and ND1 accumulate as membrane-embedded subcomplexes, whereas excess Cox4 exists as a free membrane-embedded protein in YME1L KD mitochondria. (A) YME1L KD mitochondria accumulate protein complexes containing Ndufb6 and ND1. Mitochondrial fractions were solubilized with 1% dodecyl maltoside, and equal amounts of protein extract (∼20 μg) were resolved using BN-PAGE on 8–16% polyacrylamide gradient and then immunoblotted with antibodies against Ndufb6 and ND1, the F1-α subunit of complex V, core 2 of complex III, and SDHA of complex II. Controls correspond to mitochondrial extracts from HEK293 cells transfected with the scrambled shRNA, whereas YME1L KD corresponds to mitochondrial extracts from YME1L KD cells.(B) YME1L KD mitochondria accumulate nonassembled Cox4. The immunoblots shown were prepared as in A, except that a 10–16% polyacrylamide gradient was used for electrophoretic separation and the membranes were developed with antibodies to Cox4 and Cox6c. (C) Excess Ndufb6 and Cox4 subunits accumulate as membrane-embedded polypeptides. Mitochondria (1 mg/ml) from control and YME1L KD cells were either sonicated or extracted with 100 mM sodium carbonate, pH 11.5, and centrifuged for 1 h at 144,000 × g. TCA-precipitated supernatant (S) and washed pellet (P) fractions, along with the untreated mitochondria, were immunoblotted with antibodies to ATPase F1-α, Cox1, Cox2, Cox4, and Ndufb6. (D) In contrast to Ndufb6 and ND1, only a minority of Ndufb8 is retained in complex I subcomplexes in YME1L KD cells. Immunoblots were prepared using a 5–15% polyacrylamide gradient and developed with antibodies against Ndufb6 and Ndufb8. (E) YME1L KD mitochondria exhibit increased levels of CcO subcomplexes. The immunoblots were prepared using 8–15% polyacrylamide gradient and developed with antibodies against Cox1, Cox2, and Cox5a. (F) The overexpression of Ndufb6-FLAG in HEK293 cells leads to the appearance of subcomplexes similar to those observed in YME1L KD cells. Wild-type HEK293 cells were transiently transfected with the Ndufb6-FLAG expression construct harvested at 36 h posttransfection and used to prepare mitochondrial fractions. Mitochondria were solubilized with 1% dodecyl maltoside and resolved using BN-PAGE (F) or two-dimensional BN/SDS–PAGE (G) on 5–14% polyacrylamide gels in the first dimension. Immunodetection was performed using anti-Ndufb6 and anti-FLAG antibodies. The apparent molecular weights (kDa) are estimated from the migration of complex I holoenzyme (970 kDa), complex III dimer (500 kDa), complex IV monomer (200 kDa), complex II (123 kDa), and mtHSP70 (70 kDa).
FIGURE 5:
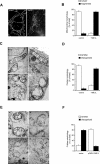
Fragmented mitochondrial network and impaired cristae morphogenesis are observed upon the loss of YME1L. (A) The loss of YME1L results in a fragmented and attenuated mitochondrial network. Mitochondria were visualized with MitoTracker Red and analyzed using a Nikon Diaphot 200 inverted microscope equipped with an Olympus DP50 camera. Images were deconvolved using the classic maximum-likelihood estimation algorithm in Huygens Professional software. Bar, 10 μM. (B) The quantification of mitochondrial network morphology in control and YME1L KD cells. Cells containing tubular (white bars) or fragmented (black bars) mitochondria were counted in a double-blind manner. More than 100 cells were scored per experiment. (C) Mitochondrial ultrastructure is abnormal in YME1L-knockdown cells. Cells were incubated in PBS containing 2% potassium permanganate for 15 min, washed with PBS, and dehydrated with an ethanol series. They were then embedded in Durcupan Epon, sectioned by microtome to thicknesses ranging from 600 to 900 Å, and stained with lead citrate and uranyl acetate. The sections were viewed with a JEOL JEM-1200 EX transmission electron microscope. Bars, 200 nM. (D) The quantification of mitochondrial cristae morphology in control and YME1L KD cells. Approximately 50 sections of individual cells were scored in a double-blind manner. Controls correspond to isolated mitochondria from HEK293 cells transfected with the scrambled shRNA.(E) Lamellar cristae morphology in YME1L-knockdown cells was restored upon the expression of wild-type YME1L. YME1L KD:: empty vector (a), YME1L KD::YME1L-FLAG (b–d). (F) The quantification of cristae morphology in YME1L KD::vector and YME1L KD::pCMV-YME1L cells. Approximately 50 sections of individual cells were scored in a double-blind manner. Error bars correspond to SD from the mean.
FIGURE 6:
Reduced growth rate and impaired mitochondrial function are observed in YME1L KD cells. (A) Growth rate is reduced in YME1L KD cells. Stable knockdown cells were seeded in six-well plates at 5 × 104 cells per well and cultured in DMEM containing 1 μg/ml puromycin. The medium was changed on the second, fourth, and sixth days. Viable cells were counted every 24 h for a total of 7 d. (B) Complex I–specific respiration was diminished in YME1L KD cells. High-resolution respirometry of digitonin-permeabilized HEK293 cells was performed with the OROBOROS oxygraph as a multiple substrate-inhibitor analysis in the presence of 0.5 μM FCCP. CI/CII, rotenone-sensitive respiration of glutamate and malate/antimycin A–sensitive respiration of succinate; CI/CIV, rotenone-sensitive respiration of glutamate and malate/sodium azide–sensitive respiration of ascorbate and TMPD; CIV/CII, sodium azide–sensitive respiration of ascorbate-TMPD and antimycin A–sensitive respiration of succinate. The loss of YME1L caused impaired apoptotic resistance. Cells were treated either with staurosporine (STS; 2 μM) for 0, 3, and 6 h (C) or H2O2 for 0 and 6 h (E), and cell lysates were analyzed by immunoblotting with cleaved PARP-specific antibody. β-Tubulin was used as a loading control. (D, F) The densitometric quantification of the cleaved PARP signal from C and E, respectively. (G) Mitochondrial proteins in YME1L KD cells exhibit increased sensitivity to oxidative damage. Cells were incubated for 3 h with 100 μM hydrogen peroxide prior to the isolation of mitochondria. Mitochondrial protein carbonylation was detected using an OxyBlot protein oxidation detection kit (Chemicon). Equal loading was verified using MemCode Reversible Protein Stain (Pierce). Signals were quantified using densitometric analysis. (H) Mitochondrial membrane proteins in YME1L KD cells exhibit increased sensitivity to oxidative damage. Cells were incubated with 200 μM hydrogen peroxide for 6 h and then used to isolate mitochondria, which were subjected to sonication and ultracentrifugation (144,000 × g; 1 h). The resulting supernatant (super) and pellet fractions were processed as in G. The signals were quantified by densitometric analysis. Controls correspond to HEK293 cells transfected with the scrambled shRNA, whereas YME1L KD corresponds to YME1L KD cells. Error bars correspond to SD from the mean. **p < 0.01, ***p < 0.001.
FIGURE 7:
The involvement of YME1L in the proteolysis of a subset of mitochondrially encoded subunits of complex I. (A) The loss of YME1L leads to the polypeptide-specific stabilization of mitochondrial translation products. Cells were labeled with a [35S]methionine-cysteine mixture in the presence of anisomycin for 1 h (pulse) and then either harvested immediately or chased for an additional 17 h (chase) in media containing unlabeled methionine prior to harvesting. The resulting lysates were subjected to 16% SDS–PAGE separation and fluorography. Equal loading was verified with Coomassie blue R-250 staining. The bottom of the fluorograph containing the ND6 signal corresponds to a longer exposure time. Controls correspond to mitochondrial extracts from HEK293 cells transfected with the scrambled shRNA, whereas YME1L KD corresponds to mitochondrial extracts from YME1L KD cells. (B) ND2, ATP6, and ND3 mRNAs are not increased in YME1L KD cells, but COX1 transcripts are significantly elevated. The relative quantification of the analyzed transcripts was performed with TaqMan Gene Expression Assays on a 7300 Real-Time PCR System. HPRT1 (hypoxanthine phosphoribosyltransferase 1) and TUBA1A (tubulin, alpha 1a) were used as reference genes. *p < 0.05, **p < 0.01. (C) The quantification of the [35S] pulse-chase experiment from A by densitometric analysis. The _y_-axis represents relative signal intensity (%).CH, chase; P, pulse. The dotted lines represent the YME1L KD sample, and the solid lines represent the controls. (D) In vivo radiolabeled ND5, ND2, Cox2, and ND6 show efficient coimmunoprecipitation with the proteolytically inactive YME1LE543Q-FLAG variant. YME1L KD cells were transiently transfected with the YME1LE543Q-FLAG construct, the wild-type YME1L construct, or the empty vector and then pulse labeled for 90 min with [35S]methionine-cysteine in the presence of emetine at 36 h posttransfection. Mitochondria were solubilized with 1% Triton X-100 and coimmunoprecipitated using Anti-FLAG M2 affinity gel (Sigma-Aldrich). The eluted antigens were separated using 16% SDS–PAGE, and radioactive signals were detected by fluorography. Black lines indicate that intervening lanes had been spliced out. The ND6 signal is partially obscured as a result of low intensity. Error bars correspond to SD from the mean.
Similar articles
- Loss of Mitochondrial AAA Proteases AFG3L2 and YME1L Impairs Mitochondrial Structure and Respiratory Chain Biogenesis.
Cesnekova J, Rodinova M, Hansikova H, Zeman J, Stiburek L. Cesnekova J, et al. Int J Mol Sci. 2018 Dec 7;19(12):3930. doi: 10.3390/ijms19123930. Int J Mol Sci. 2018. PMID: 30544562 Free PMC article. - The human homologue of the yeast mitochondrial AAA metalloprotease Yme1p complements a yeast yme1 disruptant.
Shah ZH, Hakkaart GA, Arku B, de Jong L, van der Spek H, Grivell LA, Jacobs HT. Shah ZH, et al. FEBS Lett. 2000 Aug 4;478(3):267-70. doi: 10.1016/s0014-5793(00)01859-7. FEBS Lett. 2000. PMID: 10930580 - The membrane scaffold SLP2 anchors a proteolytic hub in mitochondria containing PARL and the i-AAA protease YME1L.
Wai T, Saita S, Nolte H, Müller S, König T, Richter-Dennerlein R, Sprenger HG, Madrenas J, Mühlmeister M, Brandt U, Krüger M, Langer T. Wai T, et al. EMBO Rep. 2016 Dec;17(12):1844-1856. doi: 10.15252/embr.201642698. Epub 2016 Oct 13. EMBO Rep. 2016. PMID: 27737933 Free PMC article. - Regulation of mitochondrial plasticity by the i-AAA protease YME1L.
Ohba Y, MacVicar T, Langer T. Ohba Y, et al. Biol Chem. 2020 May 26;401(6-7):877-890. doi: 10.1515/hsz-2020-0120. Biol Chem. 2020. PMID: 32087062 Review. - ATP-dependent proteases controlling mitochondrial function in the yeast Saccharomyces cerevisiae.
Van Dyck L, Langer T. Van Dyck L, et al. Cell Mol Life Sci. 1999 Nov 30;56(9-10):825-42. doi: 10.1007/s000180050029. Cell Mol Life Sci. 1999. PMID: 11212342 Free PMC article. Review.
Cited by
- Sirt3 mitigates LPS-induced mitochondrial damage in renal tubular epithelial cells by deacetylating YME1L1.
Jian Y, Yang Y, Cheng L, Yang X, Liu H, Li W, Wan Y, Yang D. Jian Y, et al. Cell Prolif. 2023 Feb;56(2):e13362. doi: 10.1111/cpr.13362. Epub 2022 Nov 26. Cell Prolif. 2023. PMID: 36433732 Free PMC article. - The pyruvate kinase activator mitapivat reduces hemolysis and improves anemia in a β-thalassemia mouse model.
Matte A, Federti E, Kung C, Kosinski PA, Narayanaswamy R, Russo R, Federico G, Carlomagno F, Desbats MA, Salviati L, Leboeuf C, Valenti MT, Turrini F, Janin A, Yu S, Beneduce E, Ronseaux S, Iatcenko I, Dang L, Ganz T, Jung CL, Iolascon A, Brugnara C, De Franceschi L. Matte A, et al. J Clin Invest. 2021 May 17;131(10):e144206. doi: 10.1172/JCI144206. J Clin Invest. 2021. PMID: 33822774 Free PMC article. - COX-2 Expression in Hepatocytes Improves Mitochondrial Function after Hepatic Ischemia-Reperfusion Injury.
Fuertes-Agudo M, Luque-Tévar M, Cucarella C, Brea R, Boscá L, Quintana-Cabrera R, Martín-Sanz P, Casado M. Fuertes-Agudo M, et al. Antioxidants (Basel). 2022 Aug 30;11(9):1724. doi: 10.3390/antiox11091724. Antioxidants (Basel). 2022. PMID: 36139798 Free PMC article. - YME1L degradation reduces mitochondrial proteolytic capacity during oxidative stress.
Rainbolt TK, Saunders JM, Wiseman RL. Rainbolt TK, et al. EMBO Rep. 2015 Jan;16(1):97-106. doi: 10.15252/embr.201438976. Epub 2014 Nov 27. EMBO Rep. 2015. PMID: 25433032 Free PMC article. - AtOMA1 Affects the OXPHOS System and Plant Growth in Contrast to Other Newly Identified ATP-Independent Proteases in Arabidopsis Mitochondria.
Migdal I, Skibior-Blaszczyk R, Heidorn-Czarna M, Kolodziejczak M, Garbiec A, Janska H. Migdal I, et al. Front Plant Sci. 2017 Sep 7;8:1543. doi: 10.3389/fpls.2017.01543. eCollection 2017. Front Plant Sci. 2017. PMID: 28936218 Free PMC article.
References
- Augustin S, Nolden M, Muller S, Hardt O, Arnold I, Langer T. Characterization of peptides released from mitochondria: evidence for constant proteolysis and peptide efflux. J Biol Chem. 2005;280:2691–2699. - PubMed
- Benard G, Faustin B, Passerieux E, Galinier A, Rocher C, Bellance N, Delage JP, Casteilla L, Letellier T, Rossignol R. Physiological diversity of mitochondrial oxidative phosphorylation. Am J Physiol Cell Physiol. 2006;291:C1172–C1182. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases