Understanding the language of Lys36 methylation at histone H3 - PubMed (original) (raw)
Review
Understanding the language of Lys36 methylation at histone H3
Eric J Wagner et al. Nat Rev Mol Cell Biol. 2012.
Abstract
Histone side chains are post-translationally modified at multiple sites, including at Lys36 on histone H3 (H3K36). Several enzymes from yeast and humans, including the methyltransferases SET domain-containing 2 (Set2) and nuclear receptor SET domain-containing 1 (NSD1), respectively, alter the methylation status of H3K36, and significant progress has been made in understanding how they affect chromatin structure and function. Although H3K36 methylation is most commonly associated with the transcription of active euchromatin, it has also been implicated in diverse processes, including alternative splicing, dosage compensation and transcriptional repression, as well as DNA repair and recombination. Disrupted placement of methylated H3K36 within the chromatin landscape can lead to a range of human diseases, underscoring the importance of this modification.
Conflict of interest statement
Competing interests statement
The authors declare no competing financial interests.
Figures
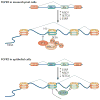
Figure 1. Domain structures of enzymes that methylate H3K36
a | A schematic of the enzymes that have been shown to promote the formation of methylated Lys36 on histone H3 (H3K36). With the exception of fly maternal-effect sterile 4 (MES-4), only human enzymes are shown. The SET domain is shown with its pre (AWS) and post domains; C5HCH is a zinc-finger (ZNF) domain; WW domains are known to interact with Pro-rich peptides; the PWWP domain is known to interact with trimethylated H3K36; and the AT hook is a DNA-binding domain. All domain assignments were derived from
Ensembl
. b | Depiction of the transitions between the multiple H3K36 methylation states, highlighting the enzymes that have been shown to function in changing a given methylation state. ASH1L, ASH1-like; BAH, bromo- associated homology; BROM, bromodomain; HMG, high mobility group; MYND, myeloid, Nervy and DEAF-1 ZNF; NIDs, nuclear receptor interaction domains; NSD, nuclear receptor SET domain-containing; PHD, plant homeodomain; RuBisCo, ribulose-1,5-bisphosphate carboxylase oxygenase; SETD, SET domain-containing; SETMAR, SET domain and mariner transposase fusion gene-containing; SMYD2, SET and MYND domain-containing 2.
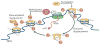
Figure 2. H3K36me3-dependent prevention of aberrant transcription in yeast
Actively transcribing RNA polymerase II (RNAPII) displaces acetylated nucleosomes. These evicted histones are reincorporated into nucleosomes and chromatin behind the polymerase. As SET domain-containing 2 (Set2) binds RNAPII, this promotes the trimethylation of Lys36 on histone H3 (H3K36me3) in the newly incorporated nucleosomes. H3K36me3 serves as a ‘mark’ that the reduced potassium dependency 3 (Rpd3) deacetylase complex binds; this complex facilitates local nucleosome deacetylation, preventing aberrant, spurious transcription in the wake of RNAPII progression through a region. CTD, carboxy-terminal domain; HAT, histone acetylase; m7G, 7-methylguanosine.
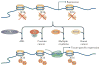
Figure 3. H3K36me3 influences alternative splicing in a cell-type specific manner
The fibroblast growth factor receptor 2 (FGFR2) locus that undergoes alternative splicing consists of two mutually exclusive exons, IIIb and IIIc, which are located between the constitutive exons 7 and 10. Mesenchymal stem cells favour the inclusion of exon IIIc and achieve this by repressing splicing of exon IIIb. Nucleosomes present near exon IIIb contain the SET domain-containing 2 (SETD2)-dependent trimethylated Lys36 on histone H3 (H3K36me3) ‘mark’ and its reader protein MORF-related gene 15 (MRG15). MRG15 also interacts with polypyrimidine tract-binding protein (PTB), a known repressor of exon inclusion, and this may be the mechanism by which the methylated H3K36 mark can influence splicing at this locus. In epithelial cells, FGFR2 expresses exon IIIb but excludes exon IIIc. Epithelial splicing regulatory protein (ESRP) is expressed and stimulates the inclusion of exon IIIb; reduced levels of H3K36me3 present at this exon, possibly as a result of lower SETD2 levels, allow its derepression. The role of dimethylases, such as the proteins of the nuclear receptor SET domain-containing (NSD) family, in this process has yet to be determined but these enzymes could also influence H3K36 methylation here.
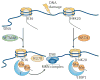
Figure 4. Model for H3K36 methylation at sites of DNA damage
Upon the generation of DNA damage, SET domain and mariner transposase fusion gene- containing (SETMAR) dimethylates Lys36 on histone H3 (H3K36) near sites of DNA double-strand breaks (DSBs), possibly by recruiting currently undefined reader proteins that facilitate the binding of KU70 and the MRE11 RAD50 NBS1 (MRN) complex to facilitate DNA repair. How methylation at H3K36 is coordinated with other chromatin modifications that are known to participate in DNA repair (such as, p53-binding protein 1 (53BP1) binding to dimethylated H4K20 (H4K20me2)) is unknown. NSD2, nuclear receptor SET domain-containing 2.
Figure 5. NSD proteins can act as oncoproteins
Through overexpression and/or translocation events that result in the fusion of nuclear receptor SET domain-containing (NSD) proteins with other proteins, such as the nucleoporin NUP98, NSD proteins can be aberrantly recruited to target loci in various tissues. As a consequence, global levels of dimethylated Lys36 on histone H3 (H3K36me2) increase and are sufficient to activate inappropriate transcription, which contributes to cancer development. In some cases, the increased levels of H3K36me2 are expected to inversely correlate with trimethylated H3K27 (H3K27me3; not shown), altering the balance between competitive activating and repressive ‘marks’. As a consequence, multiple gene sets that are sensitive to the levels of H3K36me2 are turned on, a causal event in oncogenesis. AML, acute myeloid leukaemia; AR, androgen receptor; RNAPII, RNA polymerase II.
Similar articles
- [The roles of histone lysine methylation in epigenetic regulation].
Du TT, Huang QH. Du TT, et al. Yi Chuan. 2007 Apr;29(4):387-92. doi: 10.1360/yc-007-0387. Yi Chuan. 2007. PMID: 17548299 Review. Chinese. - ASH1-catalyzed H3K36 methylation drives gene repression and marks H3K27me2/3-competent chromatin.
Bicocca VT, Ormsby T, Adhvaryu KK, Honda S, Selker EU. Bicocca VT, et al. Elife. 2018 Nov 23;7:e41497. doi: 10.7554/eLife.41497. Elife. 2018. PMID: 30468429 Free PMC article. - Shaping the cellular landscape with Set2/SETD2 methylation.
McDaniel SL, Strahl BD. McDaniel SL, et al. Cell Mol Life Sci. 2017 Sep;74(18):3317-3334. doi: 10.1007/s00018-017-2517-x. Epub 2017 Apr 6. Cell Mol Life Sci. 2017. PMID: 28386724 Free PMC article. Review. - Mixed lineage leukemia: histone H3 lysine 4 methyltransferases from yeast to human.
Malik S, Bhaumik SR. Malik S, et al. FEBS J. 2010 Apr;277(8):1805-21. doi: 10.1111/j.1742-4658.2010.07607.x. Epub 2010 Mar 4. FEBS J. 2010. PMID: 20236312 Free PMC article. Review. - Histone H3.3 G34 mutations promote aberrant PRC2 activity and drive tumor progression.
Jain SU, Khazaei S, Marchione DM, Lundgren SM, Wang X, Weinberg DN, Deshmukh S, Juretic N, Lu C, Allis CD, Garcia BA, Jabado N, Lewis PW. Jain SU, et al. Proc Natl Acad Sci U S A. 2020 Nov 3;117(44):27354-27364. doi: 10.1073/pnas.2006076117. Epub 2020 Oct 16. Proc Natl Acad Sci U S A. 2020. PMID: 33067396 Free PMC article.
Cited by
- Auditory hair cell defects as potential cause for sensorineural deafness in Wolf-Hirschhorn syndrome.
Ahmed M, Ura K, Streit A. Ahmed M, et al. Dis Model Mech. 2015 Sep;8(9):1027-35. doi: 10.1242/dmm.019547. Epub 2015 Jun 18. Dis Model Mech. 2015. PMID: 26092122 Free PMC article. - Cryo-EM structure of SETD2/Set2 methyltransferase bound to a nucleosome containing oncohistone mutations.
Liu Y, Zhang Y, Xue H, Cao M, Bai G, Mu Z, Yao Y, Sun S, Fang D, Huang J. Liu Y, et al. Cell Discov. 2021 May 11;7(1):32. doi: 10.1038/s41421-021-00261-6. Cell Discov. 2021. PMID: 33972509 Free PMC article. - Fine-tuning of chromatin composition and Polycomb recruitment by two Mi2 homologues during C. elegans early embryonic development.
Käser-Pébernard S, Pfefferli C, Aschinger C, Wicky C. Käser-Pébernard S, et al. Epigenetics Chromatin. 2016 Sep 15;9:39. doi: 10.1186/s13072-016-0091-3. eCollection 2016. Epigenetics Chromatin. 2016. PMID: 27651832 Free PMC article. - The H3K36me2 methyltransferase NSD1 modulates H3K27ac at active enhancers to safeguard gene expression.
Fang Y, Tang Y, Zhang Y, Pan Y, Jia J, Sun Z, Zeng W, Chen J, Yuan Y, Fang D. Fang Y, et al. Nucleic Acids Res. 2021 Jun 21;49(11):6281-6295. doi: 10.1093/nar/gkab473. Nucleic Acids Res. 2021. PMID: 34107030 Free PMC article. - PHRF1 promotes genome integrity by modulating non-homologous end-joining.
Chang CF, Chu PC, Wu PY, Yu MY, Lee JY, Tsai MD, Chang MS. Chang CF, et al. Cell Death Dis. 2015 Apr 9;6(4):e1716. doi: 10.1038/cddis.2015.81. Cell Death Dis. 2015. PMID: 25855964 Free PMC article.
References
- Allfrey VG, Mirsky AE. Structural modifications of histones and their possible role in the regulation of RNA Synthesis. Science. 1964;144:559. - PubMed
- Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nature Rev Genet. 2006;7:715–727. - PubMed
- Varier RA, Timmers HT. Histone lysine methylation and demethylation pathways in cancer. Biochim Biophys Acta. 2011;1815:75–89. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases