Architectural design influences the diversity and structure of the built environment microbiome - PubMed (original) (raw)
Architectural design influences the diversity and structure of the built environment microbiome
Steven W Kembel et al. ISME J. 2012 Aug.
Abstract
Buildings are complex ecosystems that house trillions of microorganisms interacting with each other, with humans and with their environment. Understanding the ecological and evolutionary processes that determine the diversity and composition of the built environment microbiome--the community of microorganisms that live indoors--is important for understanding the relationship between building design, biodiversity and human health. In this study, we used high-throughput sequencing of the bacterial 16S rRNA gene to quantify relationships between building attributes and airborne bacterial communities at a health-care facility. We quantified airborne bacterial community structure and environmental conditions in patient rooms exposed to mechanical or window ventilation and in outdoor air. The phylogenetic diversity of airborne bacterial communities was lower indoors than outdoors, and mechanically ventilated rooms contained less diverse microbial communities than did window-ventilated rooms. Bacterial communities in indoor environments contained many taxa that are absent or rare outdoors, including taxa closely related to potential human pathogens. Building attributes, specifically the source of ventilation air, airflow rates, relative humidity and temperature, were correlated with the diversity and composition of indoor bacterial communities. The relative abundance of bacteria closely related to human pathogens was higher indoors than outdoors, and higher in rooms with lower airflow rates and lower relative humidity. The observed relationship between building design and airborne bacterial diversity suggests that we can manage indoor environments, altering through building design and operation the community of microbial species that potentially colonize the human microbiome during our time indoors.
Figures
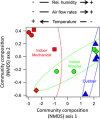
Figure 1
Ordination diagram (axis 1 and 2 from a NMDS ordination) summarizing similarity of airborne bacterial community composition (weighted UniFrac community phylogenetic dissimilarity) in samples from outdoors (blue), indoor mechanically ventilated patient rooms (red) and indoor window-ventilated patient rooms (green) at a health-care facility. Distances among communities indicate the phylogenetic similarity of bacteria in those communities. Symbols indicate sample location (○=room 229, □=room 231, ◊=room 235 and △=roof; Supplementary Table S1). Ellipses are 95% confidence intervals around samples from each environment. Arrows indicate direction of correlation between axis 1 scores from the NMDS ordination versus relative humidity (% _r_=0.67, _P_=0.01), temperature (°C; _r_=−0.68, _P_=0.01) and air flow velocity (m s−1; _r_=0.50, _P_=0.07).
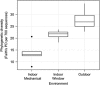
Figure 2
PD (total phylogenetic branch length; Faith's PD per 700 sequences) in different environments at a health-care facility: outdoors and indoors in patient rooms exposed to different ventilation sources (mechanical or window ventilation). PD (based on samples rarefied to 700 sequences per sample) was significantly different among all environments (Tukey's HSD; mixed model with fixed effect of environment, random effect of measurement time, overall model significant (P<0.05), pairwise differences significant (P<0.05)).
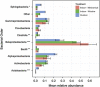
Figure 3
Taxonomic composition of airborne bacterial communities in different environments at a health-care facility: outdoors (blue) and indoors in patient rooms exposed to different ventilation sources (mechanical (red) or window (green) ventilation). Composition estimates (mean±s.d.) are based on relative abundances of bacterial 16S sequences assigned to different phyla. Asterisk symbols indicate taxonomic groups whose relative abundance differed significantly among ventilation treatments (ANOVA; *P<0.1, **P<0.05 and ***P<0.01).
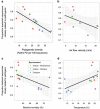
Figure 4
Relative abundance of sequences from bacterial sequences closely related to human pathogens (95% or greater sequence similarity) in airborne microbial samples versus PD and environmental conditions at a health-care facility. Solid line is best fit (with shaded 95% confidence interval) from a linear model of relative abundance of potentially pathogenic sequences versus (a) PD (total phylogenetic branch length; Faith's PD (23) per 700 sequences; % _R_2=0.53, _P_=0.005), (b) air flow velocity measured at the patient bed (m s−1; _R_2=0.27 _P_=0.04), (c) relative humidity (% _R_2=0.29, _P_=0.02) and (d) temperature (°C; _R_2=0.33, _P_=0.01).
Figure 5
Temperature and relative humidity of air in hospital patient rooms exposed to mechanical and window ventilation. Contours indicate relative abundance of sequences closely related (97% or greater sequence similarity) to the potential human pathogens (a) R. pickettii, (b) S. epidermidis and (c) S. haemolyticus, based on a polynomial spline surface fit to sample environmental coordinates.
Similar articles
- Indoor airborne bacterial communities are influenced by ventilation, occupancy, and outdoor air source.
Meadow JF, Altrichter AE, Kembel SW, Kline J, Mhuireach G, Moriyama M, Northcutt D, O'Connor TK, Womack AM, Brown GZ, Green JL, Bohannan BJ. Meadow JF, et al. Indoor Air. 2014 Feb;24(1):41-8. doi: 10.1111/ina.12047. Epub 2013 May 24. Indoor Air. 2014. PMID: 23621155 Free PMC article. - Architectural design drives the biogeography of indoor bacterial communities.
Kembel SW, Meadow JF, O'Connor TK, Mhuireach G, Northcutt D, Kline J, Moriyama M, Brown GZ, Bohannan BJ, Green JL. Kembel SW, et al. PLoS One. 2014 Jan 29;9(1):e87093. doi: 10.1371/journal.pone.0087093. eCollection 2014. PLoS One. 2014. PMID: 24489843 Free PMC article. - Relative and contextual contribution of different sources to the composition and abundance of indoor air bacteria in residences.
Miletto M, Lindow SE. Miletto M, et al. Microbiome. 2015 Dec 10;3:61. doi: 10.1186/s40168-015-0128-z. Microbiome. 2015. PMID: 26653310 Free PMC article. - The roles of the outdoors and occupants in contributing to a potential pan-microbiome of the built environment: a review.
Leung MH, Lee PK. Leung MH, et al. Microbiome. 2016 May 24;4(1):21. doi: 10.1186/s40168-016-0165-2. Microbiome. 2016. PMID: 27216717 Free PMC article. Review.
Cited by
- Significant changes in the skin microbiome mediated by the sport of roller derby.
Meadow JF, Bateman AC, Herkert KM, O'Connor TK, Green JL. Meadow JF, et al. PeerJ. 2013 Mar 12;1:e53. doi: 10.7717/peerj.53. Print 2013. PeerJ. 2013. PMID: 23638391 Free PMC article. - Freshwater Metaviromics and Bacteriophages: A Current Assessment of the State of the Art in Relation to Bioinformatic Challenges.
Bruder K, Malki K, Cooper A, Sible E, Shapiro JW, Watkins SC, Putonti C. Bruder K, et al. Evol Bioinform Online. 2016 Jun 20;12(Suppl 1):25-33. doi: 10.4137/EBO.S38549. eCollection 2016. Evol Bioinform Online. 2016. PMID: 27375355 Free PMC article. Review. - Quo vadis? Microbial profiling revealed strong effects of cleanroom maintenance and routes of contamination in indoor environments.
Moissl-Eichinger C, Auerbach AK, Probst AJ, Mahnert A, Tom L, Piceno Y, Andersen GL, Venkateswaran K, Rettberg P, Barczyk S, Pukall R, Berg G. Moissl-Eichinger C, et al. Sci Rep. 2015 Mar 17;5:9156. doi: 10.1038/srep09156. Sci Rep. 2015. PMID: 25778463 Free PMC article. - Temporal changes in the size resolved fractions of bacterial aerosols in urban and semi-urban residences.
Grydaki N, Colbeck I, Whitby C. Grydaki N, et al. Sci Rep. 2024 Aug 30;14(1):20238. doi: 10.1038/s41598-024-70495-3. Sci Rep. 2024. PMID: 39215124 Free PMC article. - Characterization of SARS-CoV-2 Distribution and Microbial Succession in a Clinical Microbiology Testing Facility during the SARS-CoV-2 Pandemic.
Sah GP, Kovalick G, Chopyk J, Kuo P, Huang L, Ghatbale P, Das P, Realegeno S, Knight R, Gilbert JA, Pride DT. Sah GP, et al. Microbiol Spectr. 2023 Mar 14;11(2):e0450922. doi: 10.1128/spectrum.04509-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36916973 Free PMC article.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
- Atkinson J, Chartier Y, Pessoa-Silva CL, Jensen P, Li Y, Seto W-H.eds). (2009Natural Ventilation for Infection Control in Health-Care Settings World Health Organization: Geneva, Switzerland - PubMed
- Badger JH, Ng PC, Venter JC.2011The human genome, microbiomes, and diseaseIn Nelson KE, (ed).Metagenomics of the Human Body Springer New York: New York, NY; 1–14.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources