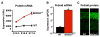Bhlhb5 and Prdm8 form a repressor complex involved in neuronal circuit assembly - PubMed (original) (raw)
. 2012 Jan 26;73(2):292-303.
doi: 10.1016/j.neuron.2011.09.035.
Alejandra E McCord, Cynthia Jung, Denize Atan, Stephanie I Mok, Martin Hemberg, Tae-Kyung Kim, John Salogiannis, Linda Hu, Sonia Cohen, Yingxi Lin, Dana Harrar, Roderick R McInnes, Michael E Greenberg
Affiliations
- PMID: 22284184
- PMCID: PMC3269007
- DOI: 10.1016/j.neuron.2011.09.035
Bhlhb5 and Prdm8 form a repressor complex involved in neuronal circuit assembly
Sarah E Ross et al. Neuron. 2012.
Abstract
Although transcription factors that repress gene expression play critical roles in nervous system development, their mechanism of action remains to be understood. Here, we report that the Olig-related transcription factor Bhlhb5 (also known as Bhlhe22) forms a repressor complex with the PR/SET domain protein, Prdm8. We find that Bhlhb5 binds to sequence-specific DNA elements and then recruits Prdm8, which mediates the repression of target genes. This interaction is critical for repressor function since mice lacking either Bhlhb5 or Prdm8 have strikingly similar cellular and behavioral phenotypes, including axonal mistargeting by neurons of the dorsal telencephalon and abnormal itch-like behavior. We provide evidence that Cadherin-11 functions as target of the Prdm8/Bhlhb5 repressor complex that must be repressed for proper neural circuit formation to occur. These findings suggest that Prdm8 is an obligate partner of Bhlhb5, forming a repressor complex that directs neural circuit assembly in part through the precise regulation of Cadherin-11.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
Figure 1. Screen for Bhlhb5 target genes identifies the PR/SET-domain protein, Prdm8
A) Affymetrix microarray-based gene profiling was performed to identify genes that are misexpressed in the dorsal telencephalon of Bhlhb5−/− mice. Three embryonic stages (E13.5, E15.5 and E17.5) were investigated, and each time point was analyzed using three independent pairs of littermate mice. Using a false discovery rate (FDR) of 0.05 or less, we identified a total of eight genes, and all of these genes were up-regulated in Bhlhb5−/− mice (also see Figures 7A and S1). Of these, Prdm8 (shown) was the most significantly misregulated. Data are mean +/− SEM, with wild type (WT) in black and Bhlhb5−/− in red, as indicated. B) Quantitative PCR confirms that Prdm8 mRNA transcripts are upregulated in the dorsal telencephalon of Bhlhb5−/− mice relative to WT littermates at E17.5. Data are normalized to WT and are presented as mean +/− SEM of biological replicates. * indicates significant difference relative to controls (p < 0.05, t-test). C) Prdm8 protein is upregulated in the cortex of Bhlhb5−/− mice at E16.5. Matched coronal sections were stained with a Prdm8 antibody raised against a GST-Prdm8 fusion protein. Also see Figure S4 for details on the generation of Prdm8 antibodies and Figure S6 for further analysis of the upregulation of Prdm8 in other regions of the nervous system and at other times during development.
Figure 2. Mice lacking either Bhlhb5 or Prdm8 have similar phenotypes
A) Absence of the corticospinal tract in the spinal cord of mice lacking either Bhlhb5 or Prdm8. In wild type mice (WT), PKCγ immunostaining labels the axons from corticospinal motor neurons as they descend in the dorsal funiculus of the spinal cord (white arrow). In both Bhlhb5−/− and Prdm8−/− mice, this axon tract is absent at all levels of the spinal cord. Similar results are observed when this fiber tract is labeled genetically (data not shown). Note that PKCγ also labels a subset of lamina II neurons in the dorsal horn, which are unaffected by the loss of Bhlhb5 or Prdm8. Images are of representative cervical spinal cords from adult mice (8 wks). B) Agenesis of the corpus callosum and hippocampal commissure in mice lacking either Bhlhb5 or Prdm8. In wild type mice (WT), dense bundles of axons connect the two hemispheres of the dorsal telencephalon (black arrow). In Bhlhb5−/− and Prdm8−/− mice, there is severe agenesis of these colossal fiber tracts. This axon targeting phenotype was observed in 20/20 Bhlhb5−/− mice, 5/7 Prdm8−/− mice, and none of the corresponding wild type littermates. Images are of representative coronal brain sections from adult mice stained with luxol fast blue and cresyl violet. C) Mice lacking either Bhlhb5−/− or Prdm8−/− show elevated scratching behavior that results in the development of skin lesions. Photos illustrating characteristic lesions are shown. All Bhlhb5−/− mice develop skin lesions at approximately 6 wks of age; ~75% of Prdm8−/− mice eventually develop skin lesions. D) Both Bhlhb5−/− and Prdm8−/− mice show an unusual movement in which they walk on their forepaws. Note that this ‘handstand’ phenotype, which appears to be secondary to abnormal contraction of the hindpaws, is only observed in a small fraction (~1– 5%) of Bhlhb5 or Prdm8 mutants. Also see Figure S2 for a more detailed analysis of axon targeting defects and Figure S3 for analysis of cortical lamination, which is unaffected by the loss of either Bhlhb5 or Prdm8.
Figure 3. Bhlhb5 and Prdm8 are colocalized in subsets of neurons
A) Double-labeling of neurons with antibodies to Bhlhb5 (green) and Prdm8 (red) reveals that Bhlhb5 and Prdm8 are localized (merge, yellow) to the nucleus where they show a diffuse pattern of expression, suggesting that they may modulate the expression level of transcribed genes found within euchromatic DNA. B) Immunohistochemistry of sagittal sections from wild type mice at P0 using antibodies to Bhlhb5 (blue) and Prdm8 (red) reveals that both proteins are co-expressed to a large extent in superficial layers of the cortical plate and in the developing hippocampus. Scale bar = 500 μm. Insets are enlarged on the right, top (i), showing co-localization (purple) of Bhlhb5 and Prdm8 in the superficial cortex, and bottom (ii), showing co-localization in sparse subsets of neurons in the diencephalon. Scale bar = 100 μm. Also see Figure S5 for a more extensive analysis of Bhlhb5 and Prdm8 co-localization at other times during development and in other regions of the nervous system.
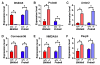
Figure 4. Mice lacking either Bhlhb5 or Prdm8 have a common molecular profile
Affymetrix microarray-based gene profiling was performed to identify genes that are misexpressed in the dorsal telencephalon of either Bhlhb5−/− or Prdm8−/− mice at P0. Each of the mutant strains was compared to its respective littermate control, using four independent biological replicates per genotype, and observations were validated by quantitative qPCR. A) Bhlhb5 mRNA is significantly upregulated in Prdm8−/− mice. B) Prdm8 mRNA is significantly upregulated in Bhlhb5−/− mice. C – F) Common genes are similarly misregulated in both Bhlhb5−/− and Prdm8−/− mice, including Antxr2 (C), Connexin36 (D), NMDA3A (E) and Paqr3 (F). Data are presented as mean +/− SEM of biological replicates and * indicates significant difference relative to controls (p < 0.05, t-test). Of note, most of the genes that are misregulated in Bhlhb5 mutant mice at P0 (i.e., as illustrated here) are different that those that are misregulated from E13.5 – E17.5 (Figure S1), suggesting that Bhlhb5 may regulate different genes at different times during development. Also see Figure S6 for analysis of possible cross-regulation of Bhlhb5 and Prdm8 at the protein level.
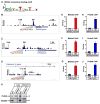
Figure 5. Bhlhb5 and Prdm8 bind to common genomic loci
DNA sequences to which Bhlhb5 is bound in the dorsal telencephalon at E17.5 were identified by chromatin immunoprecipitation followed by high throughput sequencing (ChIP-seq). A) Bhlhb5 consensus binding motif identified by the analysis of genomic Bhlhb5 binding sites. E-box is underlined. B – D) ChIP-seq data shows Bhlhb5 binding at its own proximal promoter (B), the proximal promoter of RP58 (C) and the first intron of Cdh11 (D). The height of the black bars indicates the number of input-normalized ChIP-Seq reads, representing the amount of Bhlhb5 binding. Consensus binding motifs for Bhlhb5 are found within the Bhlhb5 binding site at each gene, shown in red. Vertical gray bars indicate the genomic regions amplified in subsequent ChIP-qPCR experiments, either at the Bhlhb5 binding site (BS) or at a negative control region located −2 or −3 kb away, as indicated. The binding site for Bhlhb5 within the first intron of Cdh11 is indicated by the red bar. E – G) ChIP-qPCR using antibodies to Bhlhb5 confirms that Bhlhb5 is bound to its own proximal promoter (E), the proximal promoter of RP58 (F) and the first intron of Cdh11 (G). Experiments show significantly enriched Bhlhb5 binding at the identified Bhlhb5 binding site (BS) relative to the negative control region. H – J) ChIP-qPCR using antibodies to Prdm8 reveals that Prdm8 is also bound to the Bhlhb5 binding site at the proximal promoter of Bhlhb5 (H), the proximal promoter of RP58 (I) and the first intron of Cdh11 (J). Experiments show significantly enriched Prdm8 binding at the identified Bhlhb5 binding site (BS) relative to the negative control region. Also see Figure S7, in which 12 additional genomic loci are tested for the co-occupancy of Bhlhb5 and Prdm8. For E – J, chromatin immunoprecipitates were prepared from the dorsal telencephalon of P0 mice and y-axis (Binding) represents enrichment of gDNA over input (×10−3). For each ChIP experiment, antibody and/or knockout controls were performed in parallel, and in each case, these showed extremely low apparent binding, indicating that the ChIP experiments were specific (e.g., Figures S4C, S4D, 6A, 6B and S8). Data are representative of at least three independent experiments. * indicates significant (p < 0.05, t-test). K) Bhlhb5 and Prdm8 co-associate in neurons. Co-immunoprecipitation experiments (using ChIP conditions to preserve protein-DNA complexes) were performed to address whether Bhlhb5 and Prdm8 exist in a common complex. Bhlhb5 was immunoprecipited from the dorsal telencephalon of WT or Bhlhb5−/− mice and subjected to western blotting using antibodies to Prdm8 or Bhlhb5, as indicated. Data are representative of four independent experiments. This interaction appears to be specific since the transcriptional activator CREB was not observed in Bhlhb5-associated complexes (data not shown). Note that we were unable to co-immunoprecipitate Bhlhb5 and Prdm8 under native conditions, possibly because the interaction between Bhlhb5 and Prdm8 is indirect, or because they interact selectively in the presence of DNA.
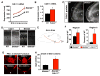
Figure 6. A Bhlhb5 dimer recruits Prdm8 to form a repressor complex at specific DNA targets
A) Bhlhb5 forms a homodimer. HEK293T cells were transfected with the indicated constructs, immunoprecipitated with antibodies to myc, and blotted using antibodies to both myc and HA, revealing that myc-tagged Bhlhb5 can co-immunoprecipitate HA-tagged Bhlhb5 but not HA-tagged E2-2. Top blot shows the immunoprecipitated protein (IP); bottom blot shows 2.5% of input. Note that myc-Bhlhb5 (with 6 myc tags) and the IgG heavy chain have the same apparent molecular weight (~ 50 kDa), whereas HA-Bhlhb5 (with 3 HA tags) is ~ 40 kDA. The isoform of E2-2 used in these experiments was the longer form, E2-2B. B) ChIP experiments performed using tissue from knockout mice reveal that Prdm8 cannot target to Bhlhb5 binding sites in the absence of Bhlhb5. In WT mice, Bhlhb5 (red) and Prdm8 (blue) bind to the proximal promoter of RP58, as revealed by ChIP-qPCR for Bhlhb5 and Prdm8 (i). In _Prdm8_−/− mice, Bhlhb5 still binds to the proximal promoter RP58 (ii). In Bhlhb5−/− mice, Prdm8 can no longer bind to Bhlhb5 binding sites, indicating that Bhlhb5 is required for Prdm8 targeting to this loci (iii). Similar results were seen for the Bhlhb5 binding sites at the Cdh11 and Bhlhb5 genes (see Figure S4). Data are representative of three independent experiments. The y-axis (Binding) represents enrichment of gDNA over input (×10−3).
Figure 7. Cdh11 is a key target of the Bhlhb5/Prdm8 repressor complex
A) Affymetrix microarray-based gene profiling identifies Cdh11 as a gene that is significantly upregulated in the dorsal telencephalon of Bhlhb5−/− mice (FDR < 0.05) from E13.5 – E17.5. Data are mean +/− SEM of biological replicates, with WT in black and Bhlhb5−/− in red, as indicated. B) Cdh11 mRNA is also upregulated in the dorsal telencephalon of Prdm8−/− mice relative to WT littermates at E14.5, as shown by qPCR. Data are normalized to WT and are presented as mean +/− SEM of biological replicates. * indicates significant difference relative to controls (p < 0.05, t-test). C–D) Cdh11 protein is upregulated in the dorsal telencephalon of Bhlhb5−/− mice. Sagittal sections from mice of the indicated genotype at E16.5 were stained with antibodies to Cdh11. Note the high Cdh11 expression in the axons of corticofugal fibers in the internal capsule (white arrows). Boxed insets are enlarged in D, which also shows the corresponding immunostaining with antibodies to Bhlhb5. E) Sagittal schematic of the brain illustrating the path of the corticospinal tract. Regions I and II (gray) indicate the location of sections in the coronal plane that were used to quantify the area of the corticospinal tract in Bhlhb5−/− mice and Bhlhb5−/−; Cdh11−/− double mutant mice (F). F) Quantification of the axon area in regions I and II. Relative to WT mice, there is a dramatic loss of axon area in the Bhlhb5−/− mice (black bars) and this loss is partially rescued in Bhlhb5−/−;Cdh11−/− double mutant mice (red bars). Five pairs of adult littermate mice were analyzed. * indicates significant (p < 0.05, t-test). G) Representative images showing the corticospinal tract stained with PKCγ in WT, _Cdh11_−/−, _Bhlhb5_−/−, and Bhlhb5−/−;Cdh11−/− mice. Matched cervical sections (corresponding to region II) from adult littermate mice are shown. The corticospinal tract is almost absent in the cervical spinal cord of Bhlhb5−/− mice but a partial rescue is seen in Bhlhb5;Cdh11−/− double mutant mice. The loss of Cdh11 alone has no effect on the formation of the corticospinal tract. H) Bhlhb5 mutant mice develop skin lesions by ~ 6 weeks of age. The onset of skin lesions is significantly delayed to ~13 weeks in Bhlhb5−/−; Cdh11−/− double mutant mice. n = 12 littermate pars; * indicates significant difference (p < 0.05, t-test.)
Figure 8. Model: Mechanism of repression
A) A Bhlhb5 homodimer binds to its consensus binding element (1) and then recruits Prdm8 to mediate repression (2). Bhlhb5 requires Prdm8 to mediate repression, whereas Prdm8 requires Bhlhb5 for proper targeting to Bhlhb5 target genes. B) A schematic illustrating phylogenetic relationships among murine Bhlhb5-related proteins and Prdm8-related proteins, including Zfp488, a zinc finger protein that shows a very high degree of similarity with Prdm8 over the C-terminus (including the C2H2 zinc fingers) but lacks the SET domain characteristic of other Prdm family members. Our study reveals that Bhlhb5 and Prdm8 are obligate partners and others have shown that Olig2 interacts physically and functionally with Zfp488 (Wang et al., 2006), suggesting that the interaction between bHLH transcription factors and Prdm-related proteins may be a general mechanism of repression during neuronal development. Note that branch lengths are not scaled to distances. C) Prdm8 and Zfp488 show a very high degree of similarity across their C-termini. Amino acids 609 – 687 (for murine Prdm8) and 258 – 337 (for murine Zfp488) are shown. Identity = 53/80 (66%); Positives = 66/80 (82%); Gaps = 1/80 (1%). E-value, 3-e28. NLS, nuclear localization sequence.
Similar articles
- Histone methyltransferase PRDM8 regulates mouse testis steroidogenesis.
Eom GH, Kim K, Kim SM, Kee HJ, Kim JY, Jin HM, Kim JR, Kim JH, Choe N, Kim KB, Lee J, Kook H, Kim N, Seo SB. Eom GH, et al. Biochem Biophys Res Commun. 2009 Oct 9;388(1):131-6. doi: 10.1016/j.bbrc.2009.07.134. Epub 2009 Jul 30. Biochem Biophys Res Commun. 2009. PMID: 19646955 - Bhlhb5 regulates the postmitotic acquisition of area identities in layers II-V of the developing neocortex.
Joshi PS, Molyneaux BJ, Feng L, Xie X, Macklis JD, Gan L. Joshi PS, et al. Neuron. 2008 Oct 23;60(2):258-72. doi: 10.1016/j.neuron.2008.08.006. Neuron. 2008. PMID: 18957218 Free PMC article. - Expression of the mouse PR domain protein Prdm8 in the developing central nervous system.
Komai T, Iwanari H, Mochizuki Y, Hamakubo T, Shinkai Y. Komai T, et al. Gene Expr Patterns. 2009 Oct;9(7):503-14. doi: 10.1016/j.gep.2009.07.005. Epub 2009 Jul 16. Gene Expr Patterns. 2009. PMID: 19616129 - Stra13 and Sharp-1, the non-grouchy regulators of development and disease.
Ow JR, Tan YH, Jin Y, Bahirvani AG, Taneja R. Ow JR, et al. Curr Top Dev Biol. 2014;110:317-38. doi: 10.1016/B978-0-12-405943-6.00009-9. Curr Top Dev Biol. 2014. PMID: 25248481 Review. - Decoding transcriptional repressor complexes in the adult central nervous system.
Adachi M, Monteggia LM. Adachi M, et al. Neuropharmacology. 2014 May;80:45-52. doi: 10.1016/j.neuropharm.2013.12.024. Epub 2014 Jan 10. Neuropharmacology. 2014. PMID: 24418103 Free PMC article. Review.
Cited by
- Reprogramming of DNA methylation at NEUROD2-bound sequences during cortical neuron differentiation.
Hahn MA, Jin SG, Li AX, Liu J, Huang Z, Wu X, Kim BW, Johnson J, Bilbao AV, Tao S, Yim JA, Fong Y, Goebbels S, Schwab MH, Lu Q, Pfeifer GP. Hahn MA, et al. Sci Adv. 2019 Oct 23;5(10):eaax0080. doi: 10.1126/sciadv.aax0080. eCollection 2019 Oct. Sci Adv. 2019. PMID: 31681843 Free PMC article. - Transcriptomic analyses of NeuroD1-mediated astrocyte-to-neuron conversion.
Ma NX, Puls B, Chen G. Ma NX, et al. Dev Neurobiol. 2022 Jul;82(5):375-391. doi: 10.1002/dneu.22882. Epub 2022 May 23. Dev Neurobiol. 2022. PMID: 35606902 Free PMC article. - N-terminal acetylation of transcription factor LIP induces immune therapy resistance via suppression of PD-L1 expression in non-small cell lung cancer.
He X, Liu Y, Gao X, Tang F, Tian Y, Gong S, Shen J, Wang A, Sun L, Wei W, Weng L. He X, et al. J Immunother Cancer. 2024 Nov 29;12(11):e009905. doi: 10.1136/jitc-2024-009905. J Immunother Cancer. 2024. PMID: 39615895 Free PMC article. - Recruitment of homodimeric proneural factors by conserved CAT-CAT E-boxes drives major epigenetic reconfiguration in cortical neurogenesis.
de Martin X, Oliva B, Santpere G. de Martin X, et al. Nucleic Acids Res. 2024 Nov 27;52(21):12895-12917. doi: 10.1093/nar/gkae950. Nucleic Acids Res. 2024. PMID: 39494521 Free PMC article. - Regulation of locomotion and motoneuron trajectory selection and targeting by the Drosophila homolog of Olig family transcription factors.
Oyallon J, Apitz H, Miguel-Aliaga I, Timofeev K, Ferreira L, Salecker I. Oyallon J, et al. Dev Biol. 2012 Sep 15;369(2):261-76. doi: 10.1016/j.ydbio.2012.06.027. Epub 2012 Jul 14. Dev Biol. 2012. PMID: 22796650 Free PMC article.
References
- Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, Surani MA. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol. 2006;8:623–630. - PubMed
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. - PubMed
- Bramblett DE, Copeland NG, Jenkins NA, Tsai MJ. BHLHB4 is a bHLH transcriptional regulator in pancreas and brain that marks the dimesencephalic boundary. Genomics. 2002;79:402–412. - PubMed
- Bramblett DE, Pennesi ME, Wu SM, Tsai MJ. The transcription factor Bhlhb4 is required for rod bipolar cell maturation. Neuron. 2004;43:779–793. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM007753/GM/NIGMS NIH HHS/United States
- R01 NS028829/NS/NINDS NIH HHS/United States
- P30 HD018655/HD/NICHD NIH HHS/United States
- P30-HD-18655/HD/NICHD NIH HHS/United States
- R37 NS028829/NS/NINDS NIH HHS/United States
- NS028829/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous