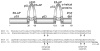Cellular transformation by human papillomaviruses: lessons learned by comparing high- and low-risk viruses - PubMed (original) (raw)
Review
Cellular transformation by human papillomaviruses: lessons learned by comparing high- and low-risk viruses
Aloysius J Klingelhutz et al. Virology. 2012.
Abstract
The oncogenic potential of papillomaviruses (PVs) has been appreciated since the 1930s yet the mechanisms of virally-mediated cellular transformation are still being revealed. Reasons for this include: a) the oncoproteins are multifunctional, b) there is an ever-growing list of cellular interacting proteins, c) more than one cellular protein may bind to a given region of the oncoprotein, and d) there is only limited information on the proteins encoded by the corresponding non-oncogenic PVs. The perspective of this review will be to contrast the activities of the viral E6 and E7 proteins encoded by the oncogenic human PVs (termed high-risk HPVs) to those encoded by their non-oncogenic counterparts (termed low-risk HPVs) in an attempt to sort out viral life cycle-related functions from oncogenic functions. The review will emphasize lessons learned from the cell culture studies of the HPVs causing mucosal/genital tract cancers.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
Fig. 1
Schematic of the HPV 16 E6 protein. The two zinc binding sites are conserved among the different HPV types. Protein binding regions are shown for HPV 16 E6. The text below the schematic displays the aligned sequences of high-risk HPV 6 E6 and low-risk HPV 16 E6.
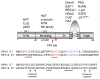
Fig. 2
Schematic of the HPV 16 E7 protein. The positions of CR1, CR2, and the zinc binding site, as well as the LXCXE motif and CKII site within CR2 are shown. The zinc binding site is conserved among the different HPV types. Protein binding regions are shown for HPV 16 E7. The text below the schematic displays the aligned sequences of high-risk HPV 6 E7 and HPV 16 E7. The PTLHE (6–10), DLYC (22–26) sequences referred to in the text are underlined; the amino acid conferring low or high Rb binding affinity is italicized; the PKC site in HPV 6 E7 is in purple; the LXCXE motif in red; and the CKII recognition sequence in blue. The proximity of the sequences required for abrogating p21CIP1 activity (aa 68–70 and 79–83) and for binding HDAC is denoted by underlining and brown lettering, respectively.
Similar articles
- The human papillomavirus oncoproteins: a review of the host pathways targeted on the road to transformation.
Scarth JA, Patterson MR, Morgan EL, Macdonald A. Scarth JA, et al. J Gen Virol. 2021 Mar;102(3):001540. doi: 10.1099/jgv.0.001540. Epub 2021 Jan 11. J Gen Virol. 2021. PMID: 33427604 Free PMC article. Review. - Human papillomavirus E6 and E7 oncoproteins as risk factors for tumorigenesis.
Ganguly N, Parihar SP. Ganguly N, et al. J Biosci. 2009 Mar;34(1):113-23. doi: 10.1007/s12038-009-0013-7. J Biosci. 2009. PMID: 19430123 Review. - The human papillomavirus family and its role in carcinogenesis.
Tommasino M. Tommasino M. Semin Cancer Biol. 2014 Jun;26:13-21. doi: 10.1016/j.semcancer.2013.11.002. Epub 2013 Dec 4. Semin Cancer Biol. 2014. PMID: 24316445 Review. - Human papillomavirus immortalization and transformation functions.
Münger K, Howley PM. Münger K, et al. Virus Res. 2002 Nov;89(2):213-28. doi: 10.1016/s0168-1702(02)00190-9. Virus Res. 2002. PMID: 12445661 Review. - Human Papillomaviruses-Associated Cancers: An Update of Current Knowledge.
Pešut E, Đukić A, Lulić L, Skelin J, Šimić I, Milutin Gašperov N, Tomaić V, Sabol I, Grce M. Pešut E, et al. Viruses. 2021 Nov 6;13(11):2234. doi: 10.3390/v13112234. Viruses. 2021. PMID: 34835040 Free PMC article. Review.
Cited by
- Molecular archeological evidence in support of the repeated loss of a papillomavirus gene.
Van Doorslaer K, McBride AA. Van Doorslaer K, et al. Sci Rep. 2016 Sep 8;6:33028. doi: 10.1038/srep33028. Sci Rep. 2016. PMID: 27604338 Free PMC article. - SKA3 promotes cell proliferation and migration in cervical cancer by activating the PI3K/Akt signaling pathway.
Hu R, Wang MQ, Niu WB, Wang YJ, Liu YY, Liu LY, Wang M, Zhong J, You HY, Wu XH, Deng N, Lu L, Wei LB. Hu R, et al. Cancer Cell Int. 2018 Nov 14;18:183. doi: 10.1186/s12935-018-0670-4. eCollection 2018. Cancer Cell Int. 2018. PMID: 30459531 Free PMC article. - Human papillomavirus E7 induces p63 expression to modulate DNA damage response.
Eldakhakhny S, Zhou Q, Crosbie EJ, Sayan BS. Eldakhakhny S, et al. Cell Death Dis. 2018 Jan 26;9(2):127. doi: 10.1038/s41419-017-0149-6. Cell Death Dis. 2018. PMID: 29374145 Free PMC article. - Oral Papillomatosis: Its Relation with Human Papilloma Virus Infection and Local Immunity-An Update.
Andrei EC, Baniță IM, Munteanu MC, Busuioc CJ, Mateescu GO, Mălin RD, Pisoschi CG. Andrei EC, et al. Medicina (Kaunas). 2022 Aug 15;58(8):1103. doi: 10.3390/medicina58081103. Medicina (Kaunas). 2022. PMID: 36013570 Free PMC article. Review. - Human papillomavirus molecular biology.
Harden ME, Munger K. Harden ME, et al. Mutat Res Rev Mutat Res. 2017 Apr-Jun;772:3-12. doi: 10.1016/j.mrrev.2016.07.002. Epub 2016 Jul 5. Mutat Res Rev Mutat Res. 2017. PMID: 28528688 Free PMC article. Review.
References
- Akgul B, Garcia-Escudero R, Ghali L, Pfister HJ, Fuchs PG, Navsaria H, Storey A. The E7 protein of cutaneous human papillomavirus type 8 causes invasion of human keratinocytes into the dermis in organotypic cultures of skin. Cancer Res. 2005;65:2216–2223. - PubMed
- Alfandari J, Shnitman Magal S, Jackman A, Schlegel R, Gonen P, Sherman L. HPV16 E6 oncoprotein inhibits apoptosis induced during serum-calcium differentiation of foreskin human keratinocytes. Virology. 1999;257:383–396. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials