Direct electron detection yields cryo-EM reconstructions at resolutions beyond 3/4 Nyquist frequency - PubMed (original) (raw)
Direct electron detection yields cryo-EM reconstructions at resolutions beyond 3/4 Nyquist frequency
Benjamin E Bammes et al. J Struct Biol. 2012 Mar.
Abstract
One limitation in electron cryo-microscopy (cryo-EM) is the inability to recover high-resolution signal from the image-recording media at the full-resolution limit of the transmission electron microscope. Direct electron detection using CMOS-based sensors for digitally recording images has the potential to alleviate this shortcoming. Here, we report a practical performance evaluation of a Direct Detection Device (DDD®) for biological cryo-EM at two different microscope voltages: 200 and 300 kV. Our DDD images of amorphous and graphitized carbon show strong per-pixel contrast with image resolution near the theoretical sampling limit of the data. Single-particle reconstructions of two frozen-hydrated bacteriophages, P22 and ε15, establish that the DDD is capable of recording usable signal for 3D reconstructions at about 4/5 of the Nyquist frequency, which is a vast improvement over the performance of conventional imaging media. We anticipate the unparalleled performance of this digital recording device will dramatically benefit cryo-EM for routine tomographic and single-particle structural determination of biological specimens.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
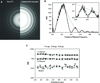
Figure 1. Graphitized carbon performance test on a JEM-2010F microscope operated at 200 kV
(A) A full frame of a graphitized carbon specimen collected at 35,700× magnification. (B) A zoomed-in view of a graphite crystal from A. (C,D) Zoomed-in views of two regions of graphite crystal from B, with lattice spacing approximately equal to the size of one pixel. (E) The raw (left) and processed (right) Fourier transform of the crystal image in B, with the characteristic graphite crystal peak visible at Nyquist frequency.
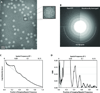
Figure 2. Carbon film performance test on a JEM-3200FSC microscope operated at 300 kV
(A) The raw (left) and processed (right) Fourier transform of an image of carbon film acquired at 10,800× detector magnification. (B) The corresponding spectral SNR curve (black) and computed SNR envelope (gray). (C) The SNR values at 1/2 Nyquist, 2/3 Nyquist, and Nyquist frequencies for carbon film images collected at 10,800× magnification at three different frame rates.
Figure 3. Evaluation of detector linearity at various detector frame rates
(A) The normalized mean intensity of flood-beam images for a wide range of total image exposures, with a zoomed view (inset) to distinguish individual data points. (B) The normalized mean intensity of flood-beam images for very low total image exposures, with a zoomed view (inset) to distinguish individual data points. (C) Spectral SNR of images of carbon film collected at 25 fps with various total image exposures. (D) The change in the spectral SNR of images of carbon film under a wide range of total image exposures.
Figure 4. ε15 bacteriophage imaging
(A) A full frame collected at 20,000× detector magnification, on a JEM-3200FSC microscope operated at 300 kV, with a zoomed-in view of a single ε15 capsid. (B) The raw (left) and processed (right) Fourier transform sum of all the particles in A with box size of 320×320 pixels. (C) The circularly averaged power spectrum computed from the Fourier transform in B. (D) The corresponding smoothed spectral SNR curve (dotted line is unsmoothed) showing distinguishable CTF oscillations close to Nyquist frequency.
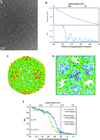
Figure 5. ε15 bacteriophage reconstructions
(A) The entire capsid reconstructed from 1380 particles, oversampled by 1.5×. (B) FSC curve of the reconstruction in A compared to a 4.5 Å reference model of ε15 (Jiang et al., 2008) for reconstructions from the original and oversampled particle images. (C) A view of the three-fold symmetry axis (black triangle) viewed from inside the reconstructed capsid with α-helices clearly visible. The backbone trace from the 4.5 Å reference model of ε15 is overlaid on our density map. (D) The FSC of reconstructions of oversampled images from 1380 and 50 particle images.
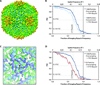
Figure 6. P22 procapsid imaging and reconstructions
(A) A full frame collected at 17,200× detector magnification acquired on a JEM-2010F microscope operating at 200 kV. (B) The circularly averaged power spectrum (top) and spectral SNR (bottom) computed from the sum of the Fourier transforms of the particle images in A. (C) 3D reconstruction of the P22 procapsid. Images were software oversampled (1.5×) prior to data processing. (D) A view of the three-fold symmetry axis (black triangle) viewed from inside the reconstructed capsid. (E) FSC curves comparing our structures using varying numbers of particle images with the previously published 3.8 Å model (Chen et al., 2011).
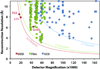
Figure 7
Single-particle reconstruction results from the EMDB (
emdatabank.org
; accessed 11/30/2011), classified by the detector type used for data collection. The three published DDD reconstructions are specifically labeled: ε15 (Fig. 5), P22 (Fig. 6), and GroEL (Milazzo et al., 2011). Note that the method for estimating resolution varies, including 0.5 FSC (square), 0.143 FSC (diamond), and other methods (circle). The solid curves show approximate resolution limits for each detector type: CCD (15 µm pixel size limited to 2/3 Nyquist frequency), film (6.35 µm limited to 2/3 Nyquist frequency), and DDD (6 µm pixel size limited to 4/5 Nyquist frequency).
Figure 8
Circularly-averaged power spectra (top) and smoothed spectral SNR curves (bottom; dotted line is unsmoothed) from low-exposure images collected at 200 kV with the following conditions: (A) Detector exposure of 10.8 e−/pixel at ~20,000× magnification, corresponding to 1.2 e−/Å2 on the specimen. (B) Detector exposure of 2.4 e−/pixel at ~60,000× magnification, corresponding to 2.4 e−/Å2 on the specimen.
Similar articles
- Practical performance evaluation of a 10k × 10k CCD for electron cryo-microscopy.
Bammes BE, Rochat RH, Jakana J, Chiu W. Bammes BE, et al. J Struct Biol. 2011 Sep;175(3):384-93. doi: 10.1016/j.jsb.2011.05.012. Epub 2011 May 17. J Struct Biol. 2011. PMID: 21619932 Free PMC article. - Observation of Bacteriophage Ultrastructure by Cryo-electron Microscopy.
Cuervo A, Carrascosa JL. Cuervo A, et al. Methods Mol Biol. 2018;1693:43-55. doi: 10.1007/978-1-4939-7395-8_5. Methods Mol Biol. 2018. PMID: 29119431 - [Progress in filters for denoising cryo-electron microscopy images].
Huang XR, Li S, Gao S. Huang XR, et al. Beijing Da Xue Xue Bao Yi Xue Ban. 2021 Mar 3;53(2):425-433. doi: 10.19723/j.issn.1671-167X.2021.02.033. Beijing Da Xue Xue Bao Yi Xue Ban. 2021. PMID: 33879921 Free PMC article. Chinese. - Routine Collection of High-Resolution cryo-EM Datasets Using 200 KV Transmission Electron Microscope.
Koh A, Khavnekar S, Yang W, Karia D, Cats D, van der Ploeg R, Grollios F, Raschdorf O, Kotecha A, Němeček D. Koh A, et al. J Vis Exp. 2022 Mar 16;(181). doi: 10.3791/63519. J Vis Exp. 2022. PMID: 35377368 Review. - Obtaining high-resolution images of biological macromolecules by using a cryo-electron microscope with a liquid-helium cooled stage.
Mitsuoka K. Mitsuoka K. Micron. 2011 Feb;42(2):100-6. doi: 10.1016/j.micron.2010.08.006. Epub 2010 Sep 8. Micron. 2011. PMID: 20869255 Review.
Cited by
- An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Wang Z, Hryc CF, Bammes B, Afonine PV, Jakana J, Chen DH, Liu X, Baker ML, Kao C, Ludtke SJ, Schmid MF, Adams PD, Chiu W. Wang Z, et al. Nat Commun. 2014 Sep 4;5:4808. doi: 10.1038/ncomms5808. Nat Commun. 2014. PMID: 25185801 Free PMC article. - Refined Cryo-EM Structure of the T4 Tail Tube: Exploring the Lowest Dose Limit.
Zheng W, Wang F, Taylor NMI, Guerrero-Ferreira RC, Leiman PG, Egelman EH. Zheng W, et al. Structure. 2017 Sep 5;25(9):1436-1441.e2. doi: 10.1016/j.str.2017.06.017. Epub 2017 Jul 27. Structure. 2017. PMID: 28757144 Free PMC article. - Single particle tomography in EMAN2.
Galaz-Montoya JG, Flanagan J, Schmid MF, Ludtke SJ. Galaz-Montoya JG, et al. J Struct Biol. 2015 Jun;190(3):279-90. doi: 10.1016/j.jsb.2015.04.016. Epub 2015 May 5. J Struct Biol. 2015. PMID: 25956334 Free PMC article. - Recent advances in retroviruses via cryo-electron microscopy.
Mak J, de Marco A. Mak J, et al. Retrovirology. 2018 Feb 23;15(1):23. doi: 10.1186/s12977-018-0405-6. Retrovirology. 2018. PMID: 29471854 Free PMC article. Review. - Single particle analysis integrated with microscopy: a high-throughput approach for reconstructing icosahedral particles.
Yan X, Cardone G, Zhang X, Zhou ZH, Baker TS. Yan X, et al. J Struct Biol. 2014 Apr;186(1):8-18. doi: 10.1016/j.jsb.2014.02.016. Epub 2014 Mar 5. J Struct Biol. 2014. PMID: 24613762 Free PMC article.
References
- Baker LA, Smith EA, Bueler SA, Rubinstein JL. The resolution dependence of optimal exposures in liquid nitrogen temperature electron cryomicroscopy of catalase crystals. J. Struct. Biol. 2010;169:431–437. - PubMed
- Booth CR, Jakana J, Chiu W. Assessing the capabilities of a 4k×4k CCD camera for electron cryo-microscopy. J. Struct. Biol. 2006;156:556–563. - PubMed
- Booth CR, Jiang W, Baker ML, Zhou ZH, Ludtke SJ, et al. A 9 Å single particle reconstruction from CCD captured images on a 200 kV electron cryo-microscope. J. Struct. Biol. 2004;147:116–127. - PubMed
Publication types
MeSH terms
Grants and funding
- P41 RR002250-26S1/RR/NCRR NIH HHS/United States
- P41RR002250/RR/NCRR NIH HHS/United States
- PN2EY016525/EY/NEI NIH HHS/United States
- P41 GM103832/GM/NIGMS NIH HHS/United States
- T32 GM008280/GM/NIGMS NIH HHS/United States
- P41 GM103832-27/GM/NIGMS NIH HHS/United States
- T15LM007093/LM/NLM NIH HHS/United States
- PN2 EY016525-08/EY/NEI NIH HHS/United States
- T15 LM007093-10S1/LM/NLM NIH HHS/United States
- P41 RR002250/RR/NCRR NIH HHS/United States
- T32 GM008280-22/GM/NIGMS NIH HHS/United States
- PN2 EY016525/EY/NEI NIH HHS/United States
- T15 LM007093/LM/NLM NIH HHS/United States
- PN2 EY016525-07/EY/NEI NIH HHS/United States
- T32 GM008280-23/GM/NIGMS NIH HHS/United States
- P41 RR002250-26/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials