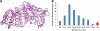Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography - PubMed (original) (raw)
Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography
Alexander Leitner et al. Mol Cell Proteomics. 2012 Mar.
Abstract
Chemical cross-linking in combination with mass spectrometric analysis offers the potential to obtain low-resolution structural information from proteins and protein complexes. Identification of peptides connected by a cross-link provides direct evidence for the physical interaction of amino acid side chains, information that can be used for computational modeling purposes. Despite impressive advances that were made in recent years, the number of experimentally observed cross-links still falls below the number of possible contacts of cross-linkable side chains within the span of the cross-linker. Here, we propose two complementary experimental strategies to expand cross-linking data sets. First, enrichment of cross-linked peptides by size exclusion chromatography selects cross-linked peptides based on their higher molecular mass, thereby depleting the majority of unmodified peptides present in proteolytic digests of cross-linked samples. Second, we demonstrate that the use of proteases in addition to trypsin, such as Asp-N, can additionally boost the number of observable cross-linking sites. The benefits of both SEC enrichment and multiprotease digests are demonstrated on a set of model proteins and the improved workflow is applied to the characterization of the 20S proteasome from rabbit and Schizosaccharomyces pombe.
Figures
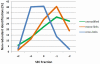
Fig. 1.
Peptide separations by size exclusion chromatography. UV traces at 215 nm are shown. A, Separation of a model peptide mixture (1 μg per peptide injected) consisting of insulin (1; 5.7 kDa), oxidized insulin A chain (2; 2.5 kDa), and angiotensin II (3; 1.0 kDa). B, Separation of the eight-protein mix cross-linked with DSS and digested with trypsin as the protease (100 μg total protein digest injected). The fractions collected for LC-MS analysis are highlighted. Elution profiles using other proteases are shown in the
supplemental Material Fig. S1
.
Fig. 2.
Relative distributions of three classes of peptides (unmodified peptides, green; mono-links, orange; and cross-links, blue) among the SEC fractions from a trypsin digest of the eight-protein mix. Data points are normalized so that for each peptide class, the sum of identifications in all five fractions is set to 100%. Distributions for other proteases are shown in
supplemental material Fig. S2
.
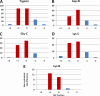
Fig. 3.
Distribution of cross-link identifications in different SEC fractions. Shown are nonredundant cross-linked peptides in five individual SEC fractions per enzyme, with the two main fractions highlighted in red. (A) Trypsin, (B) Asp-N, (C) Glu-C, (D) Lys-C, (E) Lys-N.
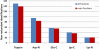
Fig. 4.
Comparison of cross-link identifications with five different proteases. Shown are nonredundant cross-linked peptides combined over all SEC fractions (in blue) and found in the two main fractions (in red), respectively.
Fig. 5.
A, Lys-Lys contacts identified by the multiprotease approach mapped onto a bovine serum albumin homology structure obtained from ModBase. Distances (Cα-Cα) of less than 30 Å, between 30 and 35 Å and above 35 Å are colored in black, orange and red, respectively. Visualization was performed using PyMOL 1.3 (Schrödinger LLC). B, Histogram showing the distribution of the BSA distance restraints shown in (A).
Similar articles
- Development of a novel cross-linking strategy for fast and accurate identification of cross-linked peptides of protein complexes.
Kao A, Chiu CL, Vellucci D, Yang Y, Patel VR, Guan S, Randall A, Baldi P, Rychnovsky SD, Huang L. Kao A, et al. Mol Cell Proteomics. 2011 Jan;10(1):M110.002212. doi: 10.1074/mcp.M110.002212. Epub 2010 Aug 24. Mol Cell Proteomics. 2011. PMID: 20736410 Free PMC article. - Probing alpha-crystallin structure using chemical cross-linkers and mass spectrometry.
Peterson JJ, Young MM, Takemoto LJ. Peterson JJ, et al. Mol Vis. 2004 Nov 16;10:857-66. Mol Vis. 2004. PMID: 15570221 - Combinatorial approach for large-scale identification of linked peptides from tandem mass spectrometry spectra.
Wang J, Anania VG, Knott J, Rush J, Lill JR, Bourne PE, Bandeira N. Wang J, et al. Mol Cell Proteomics. 2014 Apr;13(4):1128-36. doi: 10.1074/mcp.M113.035758. Epub 2014 Feb 3. Mol Cell Proteomics. 2014. PMID: 24493012 Free PMC article. - Probing structures of large protein complexes using zero-length cross-linking.
Rivera-Santiago RF, Sriswasdi S, Harper SL, Speicher DW. Rivera-Santiago RF, et al. Methods. 2015 Nov 1;89:99-111. doi: 10.1016/j.ymeth.2015.04.031. Epub 2015 May 1. Methods. 2015. PMID: 25937394 Free PMC article. Review. - Chemical cross-linking in the structural analysis of protein assemblies.
Chu F, Thornton DT, Nguyen HT. Chu F, et al. Methods. 2018 Jul 15;144:53-63. doi: 10.1016/j.ymeth.2018.05.023. Epub 2018 May 30. Methods. 2018. PMID: 29857191 Free PMC article. Review.
Cited by
- Interfaces with Structure Dynamics of the Workhorses from Cells Revealed through Cross-Linking Mass Spectrometry (CLMS).
Kalathiya U, Padariya M, Faktor J, Coyaud E, Alfaro JA, Fahraeus R, Hupp TR, Goodlett DR. Kalathiya U, et al. Biomolecules. 2021 Mar 4;11(3):382. doi: 10.3390/biom11030382. Biomolecules. 2021. PMID: 33806612 Free PMC article. Review. - Joining forces: integrating proteomics and cross-linking with the mass spectrometry of intact complexes.
Stengel F, Aebersold R, Robinson CV. Stengel F, et al. Mol Cell Proteomics. 2012 Mar;11(3):R111.014027. doi: 10.1074/mcp.R111.014027. Epub 2011 Dec 16. Mol Cell Proteomics. 2012. PMID: 22180098 Free PMC article. Review. - Advancing cell biology through proteomics in space and time (PROSPECTS).
Lamond AI, Uhlen M, Horning S, Makarov A, Robinson CV, Serrano L, Hartl FU, Baumeister W, Werenskiold AK, Andersen JS, Vorm O, Linial M, Aebersold R, Mann M. Lamond AI, et al. Mol Cell Proteomics. 2012 Mar;11(3):O112.017731. doi: 10.1074/mcp.O112.017731. Epub 2012 Feb 6. Mol Cell Proteomics. 2012. PMID: 22311636 Free PMC article. - Fast and Highly Efficient Affinity Enrichment of Azide-A-DSBSO Cross-Linked Peptides.
Matzinger M, Kandioller W, Doppler P, Heiss EH, Mechtler K. Matzinger M, et al. J Proteome Res. 2020 May 1;19(5):2071-2079. doi: 10.1021/acs.jproteome.0c00003. Epub 2020 Apr 16. J Proteome Res. 2020. PMID: 32250121 Free PMC article. - Higher-Order Structural Organization of the Mitochondrial Proteome Charted by In Situ Cross-Linking Mass Spectrometry.
Hevler JF, Heck AJR. Hevler JF, et al. Mol Cell Proteomics. 2023 Nov;22(11):100657. doi: 10.1016/j.mcpro.2023.100657. Epub 2023 Oct 6. Mol Cell Proteomics. 2023. PMID: 37805037 Free PMC article.
References
- Robinson C. V., Sali A., Baumeister W. (2007) The molecular sociology of the cell. Nature 450, 973–982 - PubMed
- Konermann L., Pan J., Liu Y. H. (2011) Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem. Soc. Rev. 40, 1224–1234 - PubMed
- Konermann L., Stocks B. B., Pan Y., Tong X. (2010) Mass spectrometry combined with oxidative labeling for exploring protein structure and folding. Mass Spectrom. Rev. 29, 651–667 - PubMed
- Benesch J. L., Ruotolo B. T., Simmons D. A., Robinson C. V. (2007) Protein complexes in the gas phase: Technology for structural genomics and proteomics. Chem. Rev. 107, 3544–3567 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources