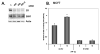Estrogenic regulation of S6K1 expression creates a positive regulatory loop in control of breast cancer cell proliferation - PubMed (original) (raw)
Estrogenic regulation of S6K1 expression creates a positive regulatory loop in control of breast cancer cell proliferation
D M Maruani et al. Oncogene. 2012.
Abstract
The 40S ribosomal S6 kinase 1 (S6K1) is an important regulator of cell growth. Expression of S6K1 is often elevated in breast cancer cells. However, the transcriptional mechanism of S6K1 overexpression is not understood. In this report, we demonstrate that estrogen activates expression of S6K1 via estrogen receptor (ER)α in ER-positive breast cancer cells. We also show that estrogen acts on the proximal promoter of the S6K1 gene in a mechanism involving the transcriptional factor GATA-3. Finally, we provide data that support the importance of estrogenic regulation of S6K1 expression in breast cancer cell proliferation. S6K1 directly phosphorylates and regulates ligand-independent activity of ERα, while ERα upregulates S6K1 expression. This S6K1-ERα relationship creates a positive feed-forward loop in control of breast cancer cell proliferation. Furthermore, the co-dependent association between S6K1 and ERα may be exploited in the development of targeted breast cancer therapies.
Conflict of interest statement
Disclosure of Potential Conflicts of Interest: None declared.
Figures
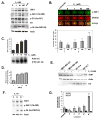
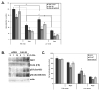
Figure 1. Estrogen upregulates S6K1 expression
(A) MCF7 cells were starved and treated with the indicated amounts of estrogen or 100nM 4-hydroxytamoxifen (4-HT). Cell lysates were immunoblotted with the indicated antibodies. (B) MCF7 cells were grown in full-serum media (untreated) or starved and treated with the indicated amounts of estrogen. In-cell western was performed by staining the cells against S6K1 (green), while DRAQ5 (red) was used as a cell normalization control. Normalized levels of S6K1 (yellow) in the merged signal image from three independent experiments are quantified in the graph, where the untreated condition is set as the 100% control. Statistical differences are shown between control and estrogen-deprived cells, as well as between estrogen-treated and estrogen-deprived cells. (C) S6K1 was immunoprecipitated from MCF7 cells treated as in (B). An in vitro kinase assay towards recombinant GST-rpS6 was performed as described in Materials and Methods, and a representative gel is shown. Levels of [γ-32P]ATP incorporation from three independent experiments were determined by autorad, quantified suing the QuantityOne software and plotted using Excel. (D) MCF7 cells were transfected with plasmids encoding ERE-firefly luciferase construct, and control Renilla luciferase. 24 h after transfection, cells were serum-starved, and treated with the indicated amounts of estrogen for an additional 24 h. Cells were lysed, and firefly luciferase expression was measured and normalized to control Renilla luciferase. The data are presented as mean ±S.D. of three independent experiments performed in triplicate. (E) Cells were grown in serum-free media for 24h, and 10nM estrogen or the ethanol vehicle was added for an additional 24h. Cell extracts were prepared, and lysates were immunoblotted with the indicated antibodies. (F) Immunoblot analysis of S6K1 levels in the mammary glands of mice subjected to estrogen treatments. (G) MCF7 cells were grown in full-serum media (untreated) or starved and stimulated with 10 nM estrogen for the indicated periods of time with or without pre-treatment with Actinomycin D. Total mRNA was isolated, and RT-qPCR analysis of S6K1 gene expression normalized to GAPDH was performed in triplicate for each sample. Statistical differences are shown between untreated and estrogen-deprived cells, as well as between estrogen-treated and estrogen-deprived cells.
Figure 2. Estrogen stimulates S6K1 promoter activity
(A) MCF7 cells were transfected with plasmids encoding RPS6KB1 promoter-regulated firefly luciferase, and control Renilla luciferase. 24 h after transfection, cells were serum-starved, and treated with the indicated amounts of estrogen for an additional 24 h. Cells were lysed, and firefly luciferase expression was measured and normalized to control Renilla luciferase. The data are presented as mean ±S.D. of each experiment performed in quadruplicate; statistical differences are calculated between estrogen-treated and estrogen deprived cells. (B) Same as in (A) with BT-474 cells. (C) ChIP analysis of promoter occupancy by GATA-3, RNA pol II phosphorylated serine 5, trimethylated histone H3 at lysine 4 and total histone H3 in MCF7 cells with or without 10 nM estrogen treatment.
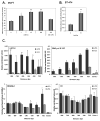
Figure 3. Overexpression of ERα stimulates RPS6KB1 promoter activity
(A) MCF7 cells were transfected with plasmids encoding ERα or empty control. 24 h after transfection, cells were incubated with serum-free media for 24 h, and treated with 10nM estrogen. Cells were lysed, and protein expression was measured by immunoblot. (B) MCF7 or HEK293E cells were transfected with a vector control or the indicated amounts of ERα-encoding plasmid. 24 h after transfection, cells were incubated in serum-free media for an additional 24 h, lysed and assayed for the indicated protein expression using immunoblot. (C) MDA-MB-231 cells were transfected with a vector control or the indicated allele of ERα-encoding plasmid. 24 h after transfection, cells were incubated in serum-free media for and additional 24 h, lysed and assayed for the indicated protein expression using immunoblot. (D) MDA-MB-231 cells were transfected with plasmids encoding ERα, RPS6KB1 promoter firefly luciferase construct, and control Renilla luciferase. 24 h after transfection, cells were serum-starved, and treated with the indicated amounts of estrogen for an additional 24 h. Cells were lysed, and firefly luciferase expression was measured and normalized to control Renilla luciferase. The data are presented as mean ±S.D. of each experiment performed in quadruplicate; statistical differences are calculated between estrogen-treated and estrogen-deprived cells. (D) Same as in (C) with HEK293E cells.
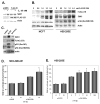
Figure 4. Down-regulation of ERα reduces S6K1 expression and abrogates estrogen-dependent RPS6KB1 promoter expression
(A) MCF7 cells were transfected with two different siRNAs targeting ERα, alone or in combination, or a scrambled control. 48 h post-transfection, cells were lysed, and protein levels in cell extracts were determined by immunoblot. (B) MCF7 cells were co- transfected with siRNA targeting ERα, the firefly luciferase RPS6KB1 reporter and Renilla luciferase control vector. 24 h after transfection, cells were serum-starved, and treated with the indicated amounts of estrogen for an additional 24 h. Cells were lysed, and firefly luciferase expression was measured and normalized to control Renilla luciferase. The data are presented as mean ±S.D. of each experiment performed in quadruplicate; statistical differences are calculated between estrogen-treated and estrogen-deprived cells.
Figure 5. ERα and S6K1 modulation regulates cell proliferation in full and low-serum media
(A) MCF7 cells were grown in full or low (1% FBS) serum media with or without rapamycin for 96 hr after siRNA transfection targeting ESR1, or siRNA knockdown together with HA-tagged S6K1 overexpression. Cell proliferation was determined using WST-1 assay. Statistical differences of three independent experiments performed in quadruplicate are calculated between the rapamycin treatment conditions and the control, as well as the siRNA knockdown versus the control or S6K1 overexpression conditions. (B) FLAG-tagged ER was overexpressed in MDA-MB-231 cells, which were subsequently grown in media with the indicated amount of serum, and assayed for protein expression by immunoblot. (C) MDA-MB-231 cells were grown in full or low (1% FBS) serum media with or without rapamycin for 96 hr after transfection of FLAG-tagged alleles of ER. Cell proliferation was determined using WST-1 assay. Statistical differences of three independent experiments performed in quadruplicate are calculated between the ER-overexpression conditions and the control.
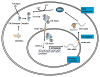
Figure 6. S6K1 and ERα participate in a positive co-regulatory relationship
ERα is activated by binding to its ligand estrogen, leading to dissociation of Heat shock proteins (HSPs), receptor dimerization, activation, and translocation to the nucleus. Phosphorylation by S6K1 promotes ligand-independent activation of ERα. In the nucleus, ERα activates the promoter region of RPS6KB1 and upregulates transcription of the S6K1 mRNA. S6K1 protein produced as a result of ERα transactivation leads to increased activation of ERα, establishing a positive feed-forward loop.
References
- Fenton TR, Gout IT. Functions and regulation of the 70kDa ribosomal S6 kinases. Int J Biochem Cell Biol. 2011 Jan;43(1):47–59. - PubMed
- Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009 May;10(5):307–318. - PubMed
- Chung J, Kuo CJ, Crabtree GR, Blenis J. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell. 1992 Jun 26;69(7):1227–1236. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous