Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation - PubMed (original) (raw)
Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation
Davis J McCarthy et al. Nucleic Acids Res. 2012 May.
Abstract
A flexible statistical framework is developed for the analysis of read counts from RNA-Seq gene expression studies. It provides the ability to analyse complex experiments involving multiple treatment conditions and blocking variables while still taking full account of biological variation. Biological variation between RNA samples is estimated separately from the technical variation associated with sequencing technologies. Novel empirical Bayes methods allow each gene to have its own specific variability, even when there are relatively few biological replicates from which to estimate such variability. The pipeline is implemented in the edgeR package of the Bioconductor project. A case study analysis of carcinoma data demonstrates the ability of generalized linear model methods (GLMs) to detect differential expression in a paired design, and even to detect tumour-specific expression changes. The case study demonstrates the need to allow for gene-specific variability, rather than assuming a common dispersion across genes or a fixed relationship between abundance and variability. Genewise dispersions de-prioritize genes with inconsistent results and allow the main analysis to focus on changes that are consistent between biological replicates. Parallel computational approaches are developed to make non-linear model fitting faster and more reliable, making the application of GLMs to genomic data more convenient and practical. Simulations demonstrate the ability of adjusted profile likelihood estimators to return accurate estimators of biological variability in complex situations. When variation is gene-specific, empirical Bayes estimators provide an advantageous compromise between the extremes of assuming common dispersion or separate genewise dispersion. The methods developed here can also be applied to count data arising from DNA-Seq applications, including ChIP-Seq for epigenetic marks and DNA methylation analyses.
Figures
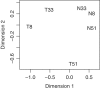
Figure 1.
Multidimensional scaling plot of the squamous cell carcinoma profiles in which distances correspond to BCV between pairs of samples. Pairwise BCVs were computed from the 500 most heterogeneous genes. Samples are labelled with patient number and either ‘T’ for tumour or ‘N’ or normal. The first plot dimension roughly corresponds to tissue source (normal or tumour) and the second to patient differences. The tumour samples are more heterogeneous than the normals.
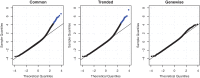
Figure 2.
QQ-plots of goodness of fit statistics using common, trended or empirical Bayes genewise (tagwise) dispersions. Genewise deviance statistics were transformed to normality, and plotted against theoretical normal quantiles. Points in blue are those genes with a significantly poor fit (Holm-adjusted _P_-value < 0.05). When using genewise dispersions, no genes show a significantly poor fit.
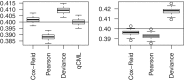
Figure 3.
Boxplots of common BCV estimates from 100 simulated data sets. The left panel shows results for the one group case, with three replicate samples in the group. The right panel shows results for a paired-design with two groups and three blocks. The horizontal lines indicate the true common BCV of 0.4, chosen to match with the carcinoma case study. Conditional maximum likelihood (qCML) is the most accurate in the former case. For generalized linear models, Cox–Reid APL is the best performer.
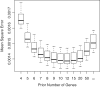
Figure 4.
Mean-square error with which empirical Bayes genewise dispersions estimate the true dispersion (BCV2), when true dispersions are randomly generated. In this case, the optimal prior weight is 10–12 prior genes, equivalent to 20–24 prior degrees of freedom. The common BCV estimator is equivalent to using infinite weight for the prior. Boxplots show results for 10 simulations.
Similar articles
- edgeR v4: powerful differential analysis of sequencing data with expanded functionality and improved support for small counts and larger datasets.
Chen Y, Chen L, Lun ATL, Baldoni PL, Smyth GK. Chen Y, et al. Nucleic Acids Res. 2025 Jan 11;53(2):gkaf018. doi: 10.1093/nar/gkaf018. Nucleic Acids Res. 2025. PMID: 39844453 Free PMC article. - No counts, no variance: allowing for loss of degrees of freedom when assessing biological variability from RNA-seq data.
Lun ATL, Smyth GK. Lun ATL, et al. Stat Appl Genet Mol Biol. 2017 Apr 25;16(2):83-93. doi: 10.1515/sagmb-2017-0010. Stat Appl Genet Mol Biol. 2017. PMID: 28599403 - BADGE: a novel Bayesian model for accurate abundance quantification and differential analysis of RNA-Seq data.
Gu J, Wang X, Halakivi-Clarke L, Clarke R, Xuan J. Gu J, et al. BMC Bioinformatics. 2014;15 Suppl 9(Suppl 9):S6. doi: 10.1186/1471-2105-15-S9-S6. Epub 2014 Sep 10. BMC Bioinformatics. 2014. PMID: 25252852 Free PMC article. - Statistical detection of differentially expressed genes based on RNA-seq: from biological to phylogenetic replicates.
Gu X. Gu X. Brief Bioinform. 2016 Mar;17(2):243-8. doi: 10.1093/bib/bbv035. Epub 2015 Jun 24. Brief Bioinform. 2016. PMID: 26108230 Review. - Genetic variation in human gene expression.
Dermitzakis ET, Stranger BE. Dermitzakis ET, et al. Mamm Genome. 2006 Jun;17(6):503-8. doi: 10.1007/s00335-006-0005-y. Epub 2006 Jun 12. Mamm Genome. 2006. PMID: 16783632 Review.
Cited by
- Extended amygdala-parabrachial circuits alter threat assessment and regulate feeding.
Luskin AT, Bhatti DL, Mulvey B, Pedersen CE, Girven KS, Oden-Brunson H, Kimbell K, Blackburn T, Sawyer A, Gereau RW 4th, Dougherty JD, Bruchas MR. Luskin AT, et al. Sci Adv. 2021 Feb 26;7(9):eabd3666. doi: 10.1126/sciadv.abd3666. Print 2021 Feb. Sci Adv. 2021. PMID: 33637526 Free PMC article. - Temporal multiomic modeling reveals a B-cell receptor proliferative program in chronic lymphocytic leukemia.
Schleiss C, Carapito R, Fornecker LM, Muller L, Paul N, Tahar O, Pichot A, Tavian M, Nicolae A, Miguet L, Mauvieux L, Herbrecht R, Cianferani S, Freund JN, Carapito C, Maumy-Bertrand M, Bahram S, Bertrand F, Vallat L. Schleiss C, et al. Leukemia. 2021 May;35(5):1463-1474. doi: 10.1038/s41375-021-01221-5. Epub 2021 Apr 8. Leukemia. 2021. PMID: 33833385 Free PMC article. - Transcriptome profiles of latently- and reactivated HIV-1 infected primary CD4+ T cells: A pooled data-analysis.
Inderbitzin A, Loosli T, Opitz L, Rusert P, Metzner KJ. Inderbitzin A, et al. Front Immunol. 2022 Aug 26;13:915805. doi: 10.3389/fimmu.2022.915805. eCollection 2022. Front Immunol. 2022. PMID: 36090997 Free PMC article. - A fourteen-lncRNA risk score system for prognostic prediction of patients with non-small cell lung cancer.
Song JY, Li XP, Qin XJ, Zhang JD, Zhao JY, Wang R. Song JY, et al. Cancer Biomark. 2020;29(4):493-508. doi: 10.3233/CBM-190505. Cancer Biomark. 2020. PMID: 32831192 Free PMC article. - An Erg-driven transcriptional program controls B cell lymphopoiesis.
Ng AP, Coughlan HD, Hediyeh-Zadeh S, Behrens K, Johanson TM, Low MSY, Bell CC, Gilan O, Chan YC, Kueh AJ, Boudier T, Feltham R, Gabrielyan A, DiRago L, Hyland CD, Ierino H, Mifsud S, Viney E, Willson T, Dawson MA, Allan RS, Herold MJ, Rogers K, Tarlinton DM, Smyth GK, Davis MJ, Nutt SL, Alexander WS. Ng AP, et al. Nat Commun. 2020 Jun 15;11(1):3013. doi: 10.1038/s41467-020-16828-y. Nat Commun. 2020. PMID: 32541654 Free PMC article.
References
- National Human Genome Research Institute (2011). DNA sequencing costs. http://www.genome.gov/sequencingcosts/
- 't Hoen PAC, Ariyurek Y, Thygesen HH, Vreugdenhil E, Vossen RHAM, Menezes RXD, Boer JM, Ommen GJBV, Dunnen JTD. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res. 2008;36:e141. - PMC - PubMed
- Mortazavi A, Williams BA, Mccue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Meth. 2008;5:621–628. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases