ATP-dependent chromatin remodeling in the DNA-damage response - PubMed (original) (raw)
ATP-dependent chromatin remodeling in the DNA-damage response
Hannes Lans et al. Epigenetics Chromatin. 2012.
Abstract
The integrity of DNA is continuously challenged by metabolism-derived and environmental genotoxic agents that cause a variety of DNA lesions, including base alterations and breaks. DNA damage interferes with vital processes such as transcription and replication, and if not repaired properly, can ultimately lead to premature aging and cancer. Multiple DNA pathways signaling for DNA repair and DNA damage collectively safeguard the integrity of DNA. Chromatin plays a pivotal role in regulating DNA-associated processes, and is itself subject to regulation by the DNA-damage response. Chromatin influences access to DNA, and often serves as a docking or signaling site for repair and signaling proteins. Its structure can be adapted by post-translational histone modifications and nucleosome remodeling, catalyzed by the activity of ATP-dependent chromatin-remodeling complexes. In recent years, accumulating evidence has suggested that ATP-dependent chromatin-remodeling complexes play important, although poorly characterized, roles in facilitating the effectiveness of the DNA-damage response. In this review, we summarize the current knowledge on the involvement of ATP-dependent chromatin remodeling in three major DNA repair pathways: nucleotide excision repair, homologous recombination, and non-homologous end-joining. This shows that a surprisingly large number of different remodeling complexes display pleiotropic functions during different stages of the DNA-damage response. Moreover, several complexes seem to have multiple functions, and are implicated in various mechanistically distinct repair pathways.
Figures
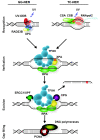
Figure 1
Mammalian nucleotide excision repair (NER). NER removes a wide variety of helix-distorting lesions from DNA, including those induced by UV light [20]. NER is executed by two different damage-detection mechanisms, which utilize the same machinery to excise and repair the damage. Transcription-coupled NER (TC-NER) is initiated by stalling of RNA polymerase 2 on a lesion present in the transcribed strand of active genes, and depends on recruitment of the CSA and CSB proteins. Other lesions are removed by global genome NER (GG-NER), which is initiated by the UV-DDB ubiquitin ligase complex and the heterotrimeric XPC/RAD23/CETN2 complex. Following detection, the transcription factor II H (TFIIH) complex is recruited, and unwinds a stretch of approximately 30 nucleotides around the damage, providing access for other repair factors. The DNA-binding proteins XPA and RPA are thought to stimulate the translocation and damage-verification activity of TFIIH and to stabilize and orient the XPF/ERCC1 and XPG endonucleases, which subsequently incise the DNA around the damage. After excision of the damaged strand, the resulting gap of 25 to 29 nucleotides is filled in by DNA synthesis and ligation, involving replication factors such as PCNA and RFC, DNA polymerases δ, ε and κ, and final sealing of the gap by DNA ligases I and III. For clarity, not all proteins known to be involved in NER are shown.
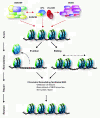
Figure 2
Mammalian nucleotide excision repair (NER)-associated chromatin remodeling. Both the SWI/SNF and the INO80 ATP-dependent chromatin-remodeling complexes are recruited to sites of UV-induced DNA damage, and are implicated in mammalian global genome NER (GG-NER). SWI/SNF may interact with the damage-detection complexes XPC/RAD23 and UV-DDB, and stimulate recruitment of XPC to the damage. Recruitment of SWI/SNF is also stimulated by XPC. In addition, mammalian INO80 interacts with UV-DDB, and stimulates recruitment of XPC. Together, SWI/SNF and INO80 are thought to regulate accessibility of DNA by sliding or eviction of nucleosomes at the damaged site. Red dotted arrows depict chromatin recruitment and protein-protein interactions. See main text for details and references.
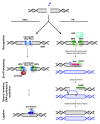
Figure 3
Mammalian double-strand break (DSB) repair. DNA DSBs are predominantly repaired by either non-homologous end-joining (NHEJ) or homologous recombination (HR) [156]. NHEJ rejoins broken DNA ends, and often requires trimming of DNA before ligation can occur. This can lead to loss of genetic information. In NHEJ, the broken DNA ends are bound by the KU70/KU80 heterodimer, which orchestrates the activity of other repair factors and recruits the phosphatidylinositol 3-kinase DNA-PKcs/PRKDC. DNA-PKcs phosphorylates and activates additional repair proteins, including itself and the ARTEMIS/DCLRE1C nuclease. ARTEMIS and/or the heterotrimeric MRE11-RAD50-NBN complex are thought to process the DNA ends prior to ligation. The DNA ends are joined by the activity of polymerases and a ligase complex consisting of XRCC4, XLF/NHEJ1 and LIG4. In contrast to NHEJ, HR is an error-free repair pathway that utilizes a sister chromatid, present only in the S- or G2-cell cycle phase, as template to repair DSBs. HR is initiated by DNA end-resection, involving the MRE11-RAD50-NBN complex and several accessory factors including nucleases. The MRE11-RAD50-NBN complex also recruits the phosphatidylinositol 3-kinase ATM, which phosphorylates histone H2AX and many other proteins involved in repair and checkpoint signaling. Single-stranded DNA generated by DNA end-resection is bound by RPA, which is subsequently replaced by RAD51. RAD51 promotes the invasion of the single-stranded DNA to a homologous double-stranded DNA template, leading to synapsis, novel DNA synthesis, strand dissolution, and repair. Many more proteins are involved in both NHEJ and HR, which are not depicted here for clarity, as they are not referred to in the main text. For details, see recent reviews by Lieber [81] and San Filippo et al. [80].
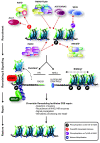
Figure 4
ATP-dependent chromatin remodeling during mammalian double-strand break (DSB) repair. A variety of ATP-dependent chromatin-remodeling complexes slide, evict, and modify nucleosomes to assist DSB repair, by increasing accessibility to DNA and facilitating DNA end-resection. Recruitment of various NHEJ, HR and signaling factors, and induction and maintenance of histone modifications at the site of damage depend on the recruitment and activity of various remodeling complexes. ALC1, which regulates DSB repair, and the NuRD complex, which promotes DSB repair and aids checkpoint activation, are both recruited in a poly(ADP ribose) polymerase (PARP)-dependent manner, which may involve poly(ADP-ribos)ylation of histones. The TRRAP/Tip60 complex localizes to DSBs to acetylate histones, and promote subsequent repair and signaling events. The ACF and/or CHRAC complexes are recruited to sites of damage, and interact with KU70 to facilitate efficient NHEJ. The WICH complex phosphorylates histone H2AX in a non--canonical manner, and stimulates efficient damage signaling and recruitment of repair factors. Finally, SWI/SNF interacts with acetylated histones H3 and H4 at the site of damage and, together with INO80, facilitates phosphorylation of histone H2AX. Red dotted arrows depict chromatin recruitment and protein-protein interactions. See main text for more detail and references. *Sliding and eviction mechanisms are probably involved in both NHEJ and HR.
Similar articles
- ISWI chromatin remodeling complexes in the DNA damage response.
Aydin ÖZ, Vermeulen W, Lans H. Aydin ÖZ, et al. Cell Cycle. 2014;13(19):3016-25. doi: 10.4161/15384101.2014.956551. Cell Cycle. 2014. PMID: 25486562 Free PMC article. - Histone post-translational modifications in DNA damage response.
Méndez-Acuña L, Di Tomaso MV, Palitti F, Martínez-López W. Méndez-Acuña L, et al. Cytogenet Genome Res. 2010;128(1-3):28-36. doi: 10.1159/000296275. Epub 2010 Apr 19. Cytogenet Genome Res. 2010. PMID: 20407219 Review. - SWI/SNF: Complex complexes in genome stability and cancer.
Ribeiro-Silva C, Vermeulen W, Lans H. Ribeiro-Silva C, et al. DNA Repair (Amst). 2019 May;77:87-95. doi: 10.1016/j.dnarep.2019.03.007. Epub 2019 Mar 15. DNA Repair (Amst). 2019. PMID: 30897376 Review. - ATP-dependent chromatin-remodeling complexes in DNA double-strand break repair: remodeling, pairing and (re)pairing.
Huang J, Liang B, Qiu J, Laurent BC. Huang J, et al. Cell Cycle. 2005 Dec;4(12):1713-5. doi: 10.4161/cc.4.12.2222. Epub 2005 Dec 5. Cell Cycle. 2005. PMID: 16294042 - Remodeling and spacing factor 1 (RSF1) deposits centromere proteins at DNA double-strand breaks to promote non-homologous end-joining.
Helfricht A, Wiegant WW, Thijssen PE, Vertegaal AC, Luijsterburg MS, van Attikum H. Helfricht A, et al. Cell Cycle. 2013 Sep 15;12(18):3070-82. doi: 10.4161/cc.26033. Epub 2013 Aug 20. Cell Cycle. 2013. PMID: 23974106 Free PMC article.
Cited by
- Chromatin dynamics during nucleotide excision repair: histones on the move.
Adam S, Polo SE. Adam S, et al. Int J Mol Sci. 2012;13(9):11895-11911. doi: 10.3390/ijms130911895. Epub 2012 Sep 19. Int J Mol Sci. 2012. PMID: 23109890 Free PMC article. Review. - Selective autophagic receptor p62 regulates the abundance of transcriptional coregulator ARIP4 during nutrient starvation.
Tsuchiya M, Isogai S, Taniguchi H, Tochio H, Shirakawa M, Morohashi KI, Hiraoka Y, Haraguchi T, Ogawa H. Tsuchiya M, et al. Sci Rep. 2015 Sep 28;5:14498. doi: 10.1038/srep14498. Sci Rep. 2015. PMID: 26412716 Free PMC article. - Human Adenovirus Infection Causes Cellular E3 Ubiquitin Ligase MKRN1 Degradation Involving the Viral Core Protein pVII.
Inturi R, Mun K, Singethan K, Schreiner S, Punga T. Inturi R, et al. J Virol. 2018 Jan 17;92(3):e01154-17. doi: 10.1128/JVI.01154-17. Print 2018 Feb 1. J Virol. 2018. PMID: 29142133 Free PMC article. - Plant-specific HDT family histone deacetylases are nucleoplasmins.
Bobde RC, Kumar A, Vasudevan D. Bobde RC, et al. Plant Cell. 2022 Nov 29;34(12):4760-4777. doi: 10.1093/plcell/koac275. Plant Cell. 2022. PMID: 36069647 Free PMC article. - UVA irradiation strengthened an interaction between UBF1/2 proteins and H4K20 di-/tri-methylation.
Stixová L, Komůrková D, Svobodová Kovaříková A, Bártová E. Stixová L, et al. Chromosome Res. 2019 Mar;27(1-2):41-55. doi: 10.1007/s10577-018-9596-x. Epub 2019 Jan 4. Chromosome Res. 2019. PMID: 30610403
References
LinkOut - more resources
Full Text Sources