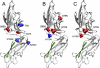Microevolution in fimH gene of mucosa-associated Escherichia coli strains isolated from pediatric patients with inflammatory bowel disease - PubMed (original) (raw)
Microevolution in fimH gene of mucosa-associated Escherichia coli strains isolated from pediatric patients with inflammatory bowel disease
Valerio Iebba et al. Infect Immun. 2012 Apr.
Abstract
Several studies reported increased numbers of mucosa-associated Escherichia coli strains in patients with inflammatory bowel disease (IBD), encompassing Crohn's disease (CD) and ulcerative colitis (UC). The majority of E. coli strains possess type 1 fimbriae, whose tip fibrillum protein, FimH, naturally undergoes amino acid replacements, an important process in the adaptation of commensal E. coli strains to environmental changes, like those observed in IBD and urinary tract infections. In this study, we analyzed mutational patterns in the fimH gene of 52 mucosa-associated E. coli strains isolated from IBD and non-IBD pediatric patients, in order to investigate microevolution of this genetic trait. FimH-positive strains were also phylogenetically typed and tested for their adhesive ability on Caco-2 cells. Specific FimH alleles for each grouping feature were found. Mutations G66S and V27A were related to CD, while mutations A242V, V163A, and T74I were attributed to UC. Otherwise, the G66S, N70S, and S78N mutations were specifically attributed to B2/D phylogroups. The N70S and A119V mutations were related to adhesive E. coli strains. Phylogroup B2, adhesive, and IBD E. coli strains showed a higher site substitution rate (SSR) in the fimH gene, together with a higher number of mutations. The degree of naïve mucosal inflammation was related to specific FimH alleles. Moreover, we could suggest that the V27A mutation is pathoadaptive for the CD intestinal habitat, while we could also suggest that both the N70S and S78N mutations are related to the more virulent E. coli B2 phylogroup. In conclusion, we found some FimH variants that seem to be more involved than others in the evolution of IBD pathogenesis.
Figures
Fig 1
Discriminating amino acid mutations in FimH protein. A three-dimensional graphical representation of the low-affinity state of FimH protein (Protein Data Bank accession number 3JWN) showed amino acid residues with the power to discriminate a patient's status (A) (red, CD; blue, UC), the E. coli phylogroup (B) (red, B2/D; blue, A/B1), and the E. coli for adhesion capability (C) (red, class 2). The donor strand of the FimG protein, which incorporates into the pilin β-barrel domain, is depicted in green.
Fig 2
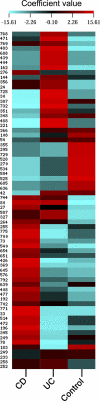
Clustered image mapping of fimH gene mutation weights. Simca-P+ software was used to compute weight coefficients for each mutation (70 x variables) on a patient's status (3 y variables) with a scaled and centered data set. These coefficients were useful to interpret the influence of the x variables on the y ones. Coefficients for different responses (y variables) were also comparable, as the y variables were normalized (scaled). As depicted in the color-coded legend, the higher that the coefficient value is, the higher the weight (red), while the lower that the value is, the lower the weight (turquoise).
Fig 3
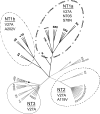
Bayesian phylogenetic tree. A Bayesian tree generated by Geneious (version 4.8.3) software as described in Materials and Methods is depicted. This rooted phylogenetic tree of the fimH sequences of 52 strains (plus the fimH sequences of strains MG1655 and LF82) showed four distinct groups: NT1a, NT1b (NT1a and NT1b were joined into NT1 in the text for SSR analysis), NT2, and NT3. These groups were progressively denoted from higher SSRs to lower ones. Inside each group, depicted with an ellipse, were reported the amino acid mutations found in ≥90% of strains belonging to the group itself. The black circle indicates the root.
Fig 4
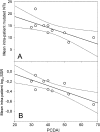
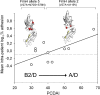
Correlation of fimH gene mutability and PCDAI. PCDAI was correlated to the mean intrapatient number of fimH mutations (A) and the mean intrapatient logarithmic SSR (B). Solid thin lines at both sides of the straight line of correlation are the 95% confidence interval.
Fig 5
Correlation of E. coli adhesion capability and PCDAI. PCDAI was correlated to the mean intrapatient logarithmic percent adhesion value. Three-dimensional FimH protein representations above the straight line of correlation depicted FimH alleles mostly found within defined PCDAI ranges. FimH allele 2 was found within strains from patients with PCDAIs ranging from 30 to 37.5, while FimH allele 5 was found within strains from patients with PCDAIs ranging from 40 to 67.5. The dashed line depicts the cutoff adhesion value of 0.8%, as described in Materials and Methods.
Similar articles
- Point mutations in FimH adhesin of Crohn's disease-associated adherent-invasive Escherichia coli enhance intestinal inflammatory response.
Dreux N, Denizot J, Martinez-Medina M, Mellmann A, Billig M, Kisiela D, Chattopadhyay S, Sokurenko E, Neut C, Gower-Rousseau C, Colombel JF, Bonnet R, Darfeuille-Michaud A, Barnich N. Dreux N, et al. PLoS Pathog. 2013 Jan;9(1):e1003141. doi: 10.1371/journal.ppat.1003141. Epub 2013 Jan 24. PLoS Pathog. 2013. PMID: 23358328 Free PMC article. - A potential role of Escherichia coli pathobionts in the pathogenesis of pediatric inflammatory bowel disease.
Schippa S, Iebba V, Totino V, Santangelo F, Lepanto M, Alessandri C, Nuti F, Viola F, Di Nardo G, Cucchiara S, Longhi C, Conte MP. Schippa S, et al. Can J Microbiol. 2012 Apr;58(4):426-32. doi: 10.1139/w2012-007. Epub 2012 Mar 22. Can J Microbiol. 2012. PMID: 22439600 - Characterization of mucosa-associated Escherichia coli strains isolated from Crohn's disease patients in Brazil.
Costa RFA, Ferrari MLA, Bringer MA, Darfeuille-Michaud A, Martins FS, Barnich N. Costa RFA, et al. BMC Microbiol. 2020 Jun 23;20(1):178. doi: 10.1186/s12866-020-01856-x. BMC Microbiol. 2020. PMID: 32576138 Free PMC article. - Intestinal colonization with phylogenetic group B2 Escherichia coli related to inflammatory bowel disease: a systematic review and meta-analysis.
Petersen AM, Halkjær SI, Gluud LL. Petersen AM, et al. Scand J Gastroenterol. 2015;50(10):1199-207. doi: 10.3109/00365521.2015.1028993. Epub 2015 Apr 24. Scand J Gastroenterol. 2015. PMID: 25910859 Review. - The potential of FimH as a novel therapeutic target for the treatment of Crohn's disease.
Sivignon A, Bouckaert J, Bernard J, Gouin SG, Barnich N. Sivignon A, et al. Expert Opin Ther Targets. 2017 Sep;21(9):837-847. doi: 10.1080/14728222.2017.1363184. Epub 2017 Aug 11. Expert Opin Ther Targets. 2017. PMID: 28762293 Review.
Cited by
- Investigation of adherent-invasive E. coli in patients with Crohn's disease.
Sarabi Asiabar A, Asadzadeh Aghdaei H, Sabokbar A, Zali MR, Feizabadi MM. Sarabi Asiabar A, et al. Med J Islam Repub Iran. 2018 Feb 11;32:11. doi: 10.14196/mjiri.32.11. eCollection 2018. Med J Islam Repub Iran. 2018. PMID: 30159262 Free PMC article. - Type 1 fimbrial phase variation in multidrug-resistant asymptomatic uropathogenic Escherichia coli clinical isolates upon adherence to HTB-4 cells.
Ghosh A, Mukherjee M. Ghosh A, et al. Folia Microbiol (Praha). 2024 Dec;69(6):1185-1204. doi: 10.1007/s12223-024-01159-y. Epub 2024 Apr 3. Folia Microbiol (Praha). 2024. PMID: 38568394 - Adhesion of Enteropathogenic, Enterotoxigenic, and Commensal Escherichia coli to the Major Zymogen Granule Membrane Glycoprotein 2.
Bartlitz C, Kolenda R, Chilimoniuk J, Grzymajło K, Rödiger S, Bauerfeind R, Ali A, Tchesnokova V, Roggenbuck D, Schierack P. Bartlitz C, et al. Appl Environ Microbiol. 2022 Mar 8;88(5):e0227921. doi: 10.1128/aem.02279-21. Epub 2022 Jan 12. Appl Environ Microbiol. 2022. PMID: 35020452 Free PMC article. - Comparative genomics reveals new single-nucleotide polymorphisms that can assist in identification of adherent-invasive Escherichia coli.
Camprubí-Font C, Lopez-Siles M, Ferrer-Guixeras M, Niubó-Carulla L, Abellà-Ametller C, Garcia-Gil LJ, Martinez-Medina M. Camprubí-Font C, et al. Sci Rep. 2018 Feb 9;8(1):2695. doi: 10.1038/s41598-018-20843-x. Sci Rep. 2018. PMID: 29426864 Free PMC article. - Genetic Diversity and Virulence Determinants of Escherichia coli Strains Isolated from Patients with Crohn's Disease in Spain and Chile.
Céspedes S, Saitz W, Del Canto F, De la Fuente M, Quera R, Hermoso M, Muñoz R, Ginard D, Khorrami S, Girón J, Assar R, Rosselló-Mora R, Vidal RM. Céspedes S, et al. Front Microbiol. 2017 May 24;8:639. doi: 10.3389/fmicb.2017.00639. eCollection 2017. Front Microbiol. 2017. PMID: 28596755 Free PMC article.
References
- Aprikian P, et al. 2007. Interdomain interaction in the FimH adhesin of Escherichia coli regulates the affinity to mannose. J. Biol. Chem. 282:23437–23446 - PubMed
- Boudeau J, Barnich N, Darfeuille-Michaud A. 2001. Type 1 pili-mediated adherence of Escherichia coli strain LF82 isolated from Crohn's disease is involved in bacterial invasion of intestinal epithelial cells. Mol. Microbiol. 39:1272–1284 - PubMed
- Bringer MA, Glasser AL, Tung CH, Méresse S, Darfeuille-Michaud A. 2006. The Crohn's disease-associated adherent-invasive Escherichia coli strain LF82 replicates in mature phagolysosomes within J774 macrophages. Cell. Microbiol. 8:471–484 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical