cn.MOPS: mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate - PubMed (original) (raw)
cn.MOPS: mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate
Günter Klambauer et al. Nucleic Acids Res. 2012 May.
Abstract
Quantitative analyses of next-generation sequencing (NGS) data, such as the detection of copy number variations (CNVs), remain challenging. Current methods detect CNVs as changes in the depth of coverage along chromosomes. Technological or genomic variations in the depth of coverage thus lead to a high false discovery rate (FDR), even upon correction for GC content. In the context of association studies between CNVs and disease, a high FDR means many false CNVs, thereby decreasing the discovery power of the study after correction for multiple testing. We propose 'Copy Number estimation by a Mixture Of PoissonS' (cn.MOPS), a data processing pipeline for CNV detection in NGS data. In contrast to previous approaches, cn.MOPS incorporates modeling of depths of coverage across samples at each genomic position. Therefore, cn.MOPS is not affected by read count variations along chromosomes. Using a Bayesian approach, cn.MOPS decomposes variations in the depth of coverage across samples into integer copy numbers and noise by means of its mixture components and Poisson distributions, respectively. The noise estimate allows for reducing the FDR by filtering out detections having high noise that are likely to be false detections. We compared cn.MOPS with the five most popular methods for CNV detection in NGS data using four benchmark datasets: (i) simulated data, (ii) NGS data from a male HapMap individual with implanted CNVs from the X chromosome, (iii) data from HapMap individuals with known CNVs, (iv) high coverage data from the 1000 Genomes Project. cn.MOPS outperformed its five competitors in terms of precision (1-FDR) and recall for both gains and losses in all benchmark data sets. The software cn.MOPS is publicly available as an R package at http://www.bioinf.jku.at/software/cnmops/ and at Bioconductor.
Figures
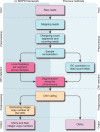
Figure 1.
The processing pipelines for CNV detection in NGS data. Left column: modeling across samples and integer copy number estimation are unique to cn.MOPS. Right column: either GC correction [class (a) methods] or read count ratios [class (b) methods] are required for previous pipelines.
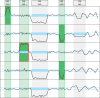
Figure 2.
Illustration of the basic concept of cn.MOPS: a CNV call incorporates the detection of variation across samples (I/NI call) and the detection of variation along a chromosome (segmentation). Curves show read counts along one chromosome for five samples. I/NI calls (green) detect variation across samples (green vertical boxes). A CNV (red box) is called if consecutive segments have high I/NI calls. Blue boxes mark segments that segmentation algorithm of class (a) methods (see the ‘Introduction’ section) would combine into a CNV. First vertical bar (from the left) and first sample: the I/NI call indicates variation across samples (‘I/NI call +’). However, too few adjacent segments show high I/NI calls. Second bar and third sample: the I/NI call indicates variation across samples (‘I/NI call +’) and sufficiently many adjacent segments show high I/NI calls, which leads to a CNV call (red box). Third bar: the read counts drop consistently and would thus be detected by a segmentation algorithm of class (a) methods (blue boxes). However, the read counts of the samples do not vary, which does not lead to an I/NI call (‘I/NI call −’). A CNV is not detected, which is correct as the copy number does not vary across samples. Fourth bar and samples numbers 2 and 4: I/NI call indicates variation across samples (‘I/NI call +’). As in the first bar, too few adjacent segments show high I/NI calls. Fifth bar and second sample: a segmentation algorithm of class (a) methods would combine adjacent read counts that are consistently small (blue box) into a CNV. However, the read counts are within the variation of the constant copy number at this location. Therefore, the I/NI call does not indicate variation across samples (‘I/NI call −’).
Figure 3.
Definitions for the evaluation of copy number detection methods. A genome is split into equally sized evaluation segments of a length shorter than the shortest CNV. Top panel: Knowing the true CNV regions (green), the evaluation segments are labeled as class 1 (CNV segment) or class −1 (non-CNV segment). Middle panel: A CNV detection method classifies each evaluation segment into CNV segments (blue, class 1) and non-CNV segments (class −1). Bottom panel: In the first line, positives (known CNV regions) are divided into true positives (TP, green) and false negatives (FN, red). In the second line, negatives (no overlap with known CNV regions) are divided into true negatives (TN, green) and false positives (FP, red). Segments partly overlapping with known or predicted CNV regions are not considered (‘na’).
Figure 4.
Whole-genome CNV calling plots that visualize the performance of cn.MOPS, MOFDOC, EWT, JointSLM, CNV-Seq, and FREEC at rediscovering known CNVs of HapMap individuals. The plots visualize CNV calling values (vertical axis) along chromosomes 1–22 of the human genome without segmentation. The first panel shows the I/NI call used for cn.MOPS. The second panel provides mean _z_-scores used by EWT, JointSLM, while the last panel depicts mean log-ratios used by CNV-Seq and FREEC. We called the largest 0.5% of the CNV calling values (blue dots) and scaled them to maximum one. Darker shades of blue indicate a high density of calling values. True CNV regions are displayed as light red bars, and the corresponding CNV calls are indicated by red dots. Segments without calling values (white segments) correspond to assembly gaps in the reference genome. A perfect calling method would call all segments in true CNV regions (red dots) at maximum 1 and would call others (blue dots) at minimum 0. Arrows indicate segments in true CNV regions that are called by one method group but not by the other method groups. A threshold of 0.6 for log-ratios-based methods, namely CNV-Seq and FREEC, and a threshold of 0.8 for cn.MOPS would lead to the same true positive rate, while cn.MOPS yields fewer false discoveries (lower FDR). cn.MOPS is better at separating segments of true CNV regions from non-CNV segments than the other methods, as indicated by the lower variance of I/NI values (see blue area at the bottom of the first panel). The better separation by cn.MOPS results in FDRs lower than those of other methods, regardless of the calling thresholds.
Figure 5.
CNV detection performance for different levels of coverage. Each curve in the two panels corresponds to the recall of one method at detecting short CNVs of lengths 1–5 kb (left panel: gains; right panel: losses). The FDR was fixed at 0.05.
Similar articles
- cn.FARMS: a latent variable model to detect copy number variations in microarray data with a low false discovery rate.
Clevert DA, Mitterecker A, Mayr A, Klambauer G, Tuefferd M, De Bondt A, Talloen W, Göhlmann H, Hochreiter S. Clevert DA, et al. Nucleic Acids Res. 2011 Jul;39(12):e79. doi: 10.1093/nar/gkr197. Epub 2011 Apr 12. Nucleic Acids Res. 2011. PMID: 21486749 Free PMC article. - Study on detection of CNVs using human whole genome bisulfite sequencing data.
Xu DT, Wang YF, Cai JL, Gong WT, Pan XC, Tian YH, Shen QP, Li JQ, Yuan XL. Xu DT, et al. Yi Chuan. 2023 Apr 20;45(4):324-340. doi: 10.16288/j.yczz.22-385. Yi Chuan. 2023. PMID: 37077166 - panelcn.MOPS: Copy-number detection in targeted NGS panel data for clinical diagnostics.
Povysil G, Tzika A, Vogt J, Haunschmid V, Messiaen L, Zschocke J, Klambauer G, Hochreiter S, Wimmer K. Povysil G, et al. Hum Mutat. 2017 Jul;38(7):889-897. doi: 10.1002/humu.23237. Epub 2017 May 16. Hum Mutat. 2017. PMID: 28449315 Free PMC article. - Free-access copy-number variant detection tools for targeted next-generation sequencing data.
Roca I, González-Castro L, Fernández H, Couce ML, Fernández-Marmiesse A. Roca I, et al. Mutat Res Rev Mutat Res. 2019 Jan-Mar;779:114-125. doi: 10.1016/j.mrrev.2019.02.005. Epub 2019 Feb 23. Mutat Res Rev Mutat Res. 2019. PMID: 31097148 Review. - Statistical challenges associated with detecting copy number variations with next-generation sequencing.
Teo SM, Pawitan Y, Ku CS, Chia KS, Salim A. Teo SM, et al. Bioinformatics. 2012 Nov 1;28(21):2711-8. doi: 10.1093/bioinformatics/bts535. Epub 2012 Aug 31. Bioinformatics. 2012. PMID: 22942022 Review.
Cited by
- DeAnnCNV: a tool for online detection and annotation of copy number variations from whole-exome sequencing data.
Zhang Y, Yu Z, Ban R, Zhang H, Iqbal F, Zhao A, Li A, Shi Q. Zhang Y, et al. Nucleic Acids Res. 2015 Jul 1;43(W1):W289-94. doi: 10.1093/nar/gkv556. Epub 2015 May 26. Nucleic Acids Res. 2015. PMID: 26013811 Free PMC article. - Next-generation sequencing is a robust strategy for the high-throughput detection of zygosity in transgenic maize.
Fritsch L, Fischer R, Wambach C, Dudek M, Schillberg S, Schröper F. Fritsch L, et al. Transgenic Res. 2015 Aug;24(4):615-23. doi: 10.1007/s11248-015-9864-x. Epub 2015 Feb 4. Transgenic Res. 2015. PMID: 25648956 - Genome-wide genetic variation discovery in Chinese Taihu pig breeds using next generation sequencing.
Wang Z, Chen Q, Liao R, Zhang Z, Zhang X, Liu X, Zhu M, Zhang W, Xue M, Yang H, Zheng Y, Wang Q, Pan Y. Wang Z, et al. Anim Genet. 2017 Feb;48(1):38-47. doi: 10.1111/age.12465. Epub 2016 Jul 27. Anim Genet. 2017. PMID: 27461929 Free PMC article. - Sister chromatid exchanges induced by perturbed replication can form independently of BRCA1, BRCA2 and RAD51.
Heijink AM, Stok C, Porubsky D, Manolika EM, de Kanter JK, Kok YP, Everts M, de Boer HR, Audrey A, Bakker FJ, Wierenga E, Tijsterman M, Guryev V, Spierings DCJ, Knipscheer P, van Boxtel R, Ray Chaudhuri A, Lansdorp PM, van Vugt MATM. Heijink AM, et al. Nat Commun. 2022 Nov 7;13(1):6722. doi: 10.1038/s41467-022-34519-8. Nat Commun. 2022. PMID: 36344511 Free PMC article. - Detection of Genomic Structural Variants from Next-Generation Sequencing Data.
Tattini L, D'Aurizio R, Magi A. Tattini L, et al. Front Bioeng Biotechnol. 2015 Jun 25;3:92. doi: 10.3389/fbioe.2015.00092. eCollection 2015. Front Bioeng Biotechnol. 2015. PMID: 26161383 Free PMC article. Review.
References
- Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470:187–197. - PubMed
- Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous