Determinants of murein hydrolase targeting to cross-wall of Staphylococcus aureus peptidoglycan - PubMed (original) (raw)
Determinants of murein hydrolase targeting to cross-wall of Staphylococcus aureus peptidoglycan
Matthew B Frankel et al. J Biol Chem. 2012.
Abstract
Cells of eukaryotic or prokaryotic origin express proteins with LysM domains that associate with the cell wall envelope of bacteria. The molecular properties that enable LysM domains to interact with microbial cell walls are not yet established. Staphylococcus aureus, a spherical microbe, secretes two murein hydrolases with LysM domains, Sle1 and LytN. We show here that the LysM domains of Sle1 and LytN direct murein hydrolases to the staphylococcal envelope in the vicinity of the cross-wall, the mid-cell compartment for peptidoglycan synthesis. LysM domains associate with the repeating disaccharide β-N-acetylmuramic acid, (1→4)-β-N-acetylglucosamine of staphylococcal peptidoglycan. Modification of N-acetylmuramic acid with wall teichoic acid, a ribitol-phosphate polymer tethered to murein linkage units, prevents the LysM domain from binding to peptidoglycan. The localization of LytN and Sle1 to the cross-wall is abolished in staphylococcal tagO mutants, which are defective for wall teichoic acid synthesis. We propose a model whereby the LysM domain ensures septal localization of LytN and Sle1 followed by processive cleavage of peptidoglycan, thereby exposing new LysM binding sites in the cross-wall and separating bacterial cells.
Figures
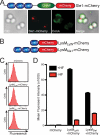
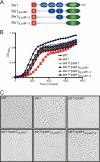
FIGURE 1.
LysM and CHAPS domains are both required for LytN localization to the cross-wall. A, diagram of the LytN-mCherry hybrid and its variants. Color-coded domains identify the LysM domain (blue), the CHAP domain (green), and the mCherry fusion (red). B, S. aureus Newman cells were fixed and incubated with purified mCherry, LytN-mCherry, LytNΔLysM-mCherry, or LytNΔCHAP-mCherry. Binding of proteins to the staphylococcal envelope was assessed by fluorescence cytometry. C, microscopy of S. aureus Newman stained with ConA and incubated with LytN-mCherry. The top left panel displays the DIC image of staphylococcal cells, which were analyzed by fluorescence microscopy (bottom panels) for red (LytN-mCherry) and green (ConA) signals. The top right panel displays a merged image derived from all three data sets. D, purified mCherry, LytN-mCherry, LytNΔLysM-mCherry, or LytNΔCHAP-mCherry was incubated with purified peptidoglycan from S. aureus Newman, which had been isolated by glass bead disruption of murein sacculi, treated with protease, and extracted with detergent, and finally WTA was removed with HF. Protein binding to peptidoglycan was analyzed with a co-sedimentation assay and fluorescence intensity measurements. All binding assays were performed in triplicate; average data and S.E. (error bars) were recorded.
FIGURE 2.
The LysM domains of LytN and Sle1 bind to staphylococcal peptidoglycan. A, purified Sle1-mCherry (see top panel diagram for the domain structure of the hybrid) was incubated with S. aureus Newman cells that had been stained with ConA. The left panel displays the DIC image of staphylococcal cells analyzed by fluorescence microscopy (middle panels) for red (Sle1-mCherry) and green (ConA) signals. The right panel displays a merged image derived from all three data sets. B, diagram for the domain structures of the LysM domains from LytN and Sle1 fused to mCherry. C, mCherry, LysMLytN-mCherry, or LysMSle1-mCherry was incubated with wild-type staphylococci and assessed for binding with fluorescence cytometry. D, mCherry, LysMLytN-mCherry, or LysMSle1-mCherry was incubated with purified peptidoglycan from S. aureus Newman, which had been isolated by glass bead disruption of murein sacculi, treated with protease, and extracted with detergent. Murein sacculi were either treated with hydrofluoric acid (+HF) or left untreated (−HF) prior to analyzing mCherry hybrids by co-sedimentation and fluorescence intensity measurements. All binding assays were performed in triplicate; average data and S.E. (error bars) were recorded.
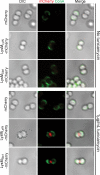
FIGURE 3.
Wall teichoic acid modification interferes with the binding of LysM domains to peptidoglycan. A, purified mCherry, LysMLytN-mCherry, or LysMSle1-mCherry was incubated with Δ_tagO_ mutant staphylococci, and binding to the bacterial envelope was measured by fluorescence cytometry. B, purified mCherry, LysMLytN-mCherry, or LysMSle1-mCherry was incubated with Δ_tagO_ mutant staphylococci that had been stained with BODIPY-vancomycin (Vancomycin). The left panels display the DIC image of staphylococcal cells analyzed by fluorescence microscopy (middle panels) for red (Cherry, LysMLytN-mCherry, or LysMSle1-mCherry) and green (vancomycin) signals. The right panels display merged images derived from all three data sets. C, Cherry, LysMLytN-mCherry, or LysMSle1-mCherry was incubated with purified peptidoglycan from Δ_tagO_ mutant staphylococci, which had been isolated by glass bead disruption of murein sacculi, treated with protease, and extracted with detergent. Murein sacculi were either treated with hydrofluoric acid (+HF) or left untreated (−HF) prior to analyzing mCherry hybrids by co-sedimentation and fluorescence intensity measurements. All binding assays were performed in triplicate; average data and S.E. (error bars) were recorded.
FIGURE 4.
The LysM domains of LytN and Sle1 bind to the cross-wall of tunicamycin-treated staphylococci. Staphylococci were grown to logarithmic phase and treated with 1 μg/ml tunicamycin (bottom panels) or left untreated (top panels) for 30 min. Staphylococci were fixed, stained with ConA, and incubated with mCherry, LysMLytN-mCherry, or LysMSle1-mCherry recombinant proteins. Cells were then adhered to a coverslip and viewed by microscopy. Left panels display the DIC image of staphylococcal cells analyzed by fluorescence microscopy (middle panels) for red (Cherry, LysMLytN-mCherry, or LysMSle1-mCherry) and green (ConA) signals. The right panels display merged images derived from all data sets.
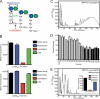
FIGURE 5.
LysM domains of LytN and Sle1 bind to the repeating disaccharide strands of staphylococcal peptidoglycan. A, diagram of the structure of S. aureus peptidoglycan, identifying the cleavage sites for mutanolysin, lysostaphin, and LytN. B, HF-treated, purified peptidoglycan was digested with mutanolysin, lysostaphin, or LytN. Soluble muropeptides were incubated with LysMLytN-mCherry or LysMSle1-mCherry. Inhibition of the binding of mCherry hybrids to staphylococcal murein sacculi was measured with co-sedimentation and fluorescence intensity measurements. All binding assays were performed in triplicate; average data and S.E. were recorded. C, RP-HPLC chromatography of lysostaphin-treated S. aureus Newman (NM) PG. D, PG compounds in the fractions of the RP-HPLC experiment from C were dissolved in water and incubated with LysMLytN-mCherry. Inhibition of the binding of LysMLytN-mCherry to staphylococcal murein sacculi was measured with co-sedimentation and fluorescence intensity measurements. Gray bars, reduction of >40%; black bars, reduction of >50%. E, lysostaphin-treated peptidoglycan was incubated with mutanolysin and subjected to RP-HPLC. Inset, binding of LysMLytN-mCherry preincubated with buffer control or lysostaphin-solubilized or lysostaphin and mutanolysin-solubilized peptidoglycan to staphylococcal murein sacculi. All binding assays were performed in triplicate; average data and S.E. (error bars) were recorded.
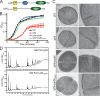
FIGURE 6.
Contribution of the LysM domain to LytN function. A, diagram of the domain structure of LytN and its LytNΔLysM variant. B, the growth of S. aureus Newman (WT) or its lytN variant with or without p_lytN_ or p_lytN_Δ_LysM_ plasmids was measured as the increase in the absorbance at 600-nm light. C, staphylococcal strains were fixed, thin sectioned, and viewed by transmission electron microscopy to visualize the cell wall envelope and cross-wall. Images of the panels on the left display a single cell; images of the panels on the right show the cross-walls of a staphylococcal cell. D, purified LytN and LytNΔLysM were incubated with highly purified staphylococcal peptidoglycan. Soluble muropeptides were separated by RP-HPLC, and muropeptides were detected by absorbance at 206-nm light. Standard error (S.E.) of the mean recorded (error bars).
FIGURE 7.
Contribution of the LysM domains to Sle1 function. A, diagram of the structural domains for Sle1 and its expression constructs with in-frame deletions of one, two, or all three LysM domains encoded by plasmid p_sle1_ or p_sle1_Δ_LysM1_, p_sle1_Δ_LysM1-2_, and p_sle1_Δ_LysM1–3. B_, growth of S. aureus Newman (WT) or its sle1 variant with or without p_sle1_ or p_sle1_Δ_LysM1_, p_sle1_Δ_LysM1-2_, and p_sle1_Δ_LysM1–3_ plasmids was measured as the increase in the absorbance at 600-nm light. C, stationary phase aliquots of staphylococcal cultures analyzed in B were viewed by light microscopy and DIC images were captured. Error bars, S.E.
Similar articles
- LytN, a murein hydrolase in the cross-wall compartment of Staphylococcus aureus, is involved in proper bacterial growth and envelope assembly.
Frankel MB, Hendrickx AP, Missiakas DM, Schneewind O. Frankel MB, et al. J Biol Chem. 2011 Sep 16;286(37):32593-605. doi: 10.1074/jbc.M111.258863. Epub 2011 Jul 22. J Biol Chem. 2011. PMID: 21784864 Free PMC article. - EssH Peptidoglycan Hydrolase Enables Staphylococcus aureus Type VII Secretion across the Bacterial Cell Wall Envelope.
Bobrovskyy M, Willing SE, Schneewind O, Missiakas D. Bobrovskyy M, et al. J Bacteriol. 2018 Sep 24;200(20):e00268-18. doi: 10.1128/JB.00268-18. Print 2018 Oct 15. J Bacteriol. 2018. PMID: 30082459 Free PMC article. - Identification and molecular characterization of an N-acetylmuramyl-L-alanine amidase Sle1 involved in cell separation of Staphylococcus aureus.
Kajimura J, Fujiwara T, Yamada S, Suzawa Y, Nishida T, Oyamada Y, Hayashi I, Yamagishi J, Komatsuzawa H, Sugai M. Kajimura J, et al. Mol Microbiol. 2005 Nov;58(4):1087-101. doi: 10.1111/j.1365-2958.2005.04881.x. Mol Microbiol. 2005. PMID: 16262792 - Murein and pseudomurein cell wall binding domains of bacteria and archaea--a comparative view.
Visweswaran GR, Dijkstra BW, Kok J. Visweswaran GR, et al. Appl Microbiol Biotechnol. 2011 Dec;92(5):921-8. doi: 10.1007/s00253-011-3637-0. Epub 2011 Oct 20. Appl Microbiol Biotechnol. 2011. PMID: 22012341 Free PMC article. Review. - Uncovering the activities, biological roles, and regulation of bacterial cell wall hydrolases and tailoring enzymes.
Do T, Page JE, Walker S. Do T, et al. J Biol Chem. 2020 Mar 6;295(10):3347-3361. doi: 10.1074/jbc.REV119.010155. Epub 2020 Jan 23. J Biol Chem. 2020. PMID: 31974163 Free PMC article. Review.
Cited by
- Sortases, Surface Proteins, and Their Roles in Staphylococcus aureus Disease and Vaccine Development.
Schneewind O, Missiakas D. Schneewind O, et al. Microbiol Spectr. 2019 Jan;7(1):10.1128/microbiolspec.psib-0004-2018. doi: 10.1128/microbiolspec.PSIB-0004-2018. Microbiol Spectr. 2019. PMID: 30737913 Free PMC article. - Noninvasive Analysis of Peptidoglycan from Living Animals.
Ocius KL, Kolli SH, Ahmad SS, Dressler JM, Chordia MD, Jutras BL, Rutkowski MR, Pires MM. Ocius KL, et al. Bioconjug Chem. 2024 Apr 17;35(4):489-498. doi: 10.1021/acs.bioconjchem.4c00007. Epub 2024 Apr 9. Bioconjug Chem. 2024. PMID: 38591251 Free PMC article. - Cell-Wall Hydrolases as Antimicrobials against Staphylococcus Species: Focus on Sle1.
Vermassen A, Talon R, Andant C, Provot C, Desvaux M, Leroy S. Vermassen A, et al. Microorganisms. 2019 Nov 12;7(11):559. doi: 10.3390/microorganisms7110559. Microorganisms. 2019. PMID: 31726796 Free PMC article. - Surfaceome and Proteosurfaceome in Parietal Monoderm Bacteria: Focus on Protein Cell-Surface Display.
Desvaux M, Candela T, Serror P. Desvaux M, et al. Front Microbiol. 2018 Feb 14;9:100. doi: 10.3389/fmicb.2018.00100. eCollection 2018. Front Microbiol. 2018. PMID: 29491848 Free PMC article. Review. - Peptidoglycan Contribution to the B Cell Superantigen Activity of Staphylococcal Protein A.
Shi M, Willing SE, Kim HK, Schneewind O, Missiakas D. Shi M, et al. mBio. 2021 Apr 20;12(2):e00039-21. doi: 10.1128/mBio.00039-21. mBio. 2021. PMID: 33879590 Free PMC article.
References
- Lowy F. D. (1998) Staphylococcus aureus infections. New Engl. J. Med. 339, 520–532 - PubMed
- Projan S. J., Nesin M., Dunman P. M. (2006) Staphylococcal vaccines and immunotherapy. To dream the impossible dream? Curr. Opin. Pharmacol. 6, 473–479 - PubMed
- Strominger J. L., Izaki K., Matsuhashi M., Tipper D. J. (1967) Peptidoglycan transpeptidase and d-alanine carboxypeptidase. Penicillin-sensitive enzymatic reactions. Fed. Proc. 26, 9–22 - PubMed
- Schneewind O., Fowler A., Faull K. F. (1995) Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science 268, 103–106 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases