Age-related somatic structural changes in the nuclear genome of human blood cells - PubMed (original) (raw)
. 2012 Feb 10;90(2):217-28.
doi: 10.1016/j.ajhg.2011.12.009. Epub 2012 Feb 2.
Chiara Rasi, Hamid R Razzaghian, Geeta Pakalapati, Lindsay Waite, Krista Stanton Thilbeault, Anna Ronowicz, Nathan E Wineinger, Hemant K Tiwari, Dorret Boomsma, Maxwell P Westerman, Jennifer R Harris, Robert Lyle, Magnus Essand, Fredrik Eriksson, Themistocles L Assimes, Carlos Iribarren, Eric Strachan, Terrance P O'Hanlon, Lisa G Rider, Frederick W Miller, Vilmantas Giedraitis, Lars Lannfelt, Martin Ingelsson, Arkadiusz Piotrowski, Nancy L Pedersen, Devin Absher, Jan P Dumanski
Affiliations
- PMID: 22305530
- PMCID: PMC3276669
- DOI: 10.1016/j.ajhg.2011.12.009
Age-related somatic structural changes in the nuclear genome of human blood cells
Lars A Forsberg et al. Am J Hum Genet. 2012.
Abstract
Structural variations are among the most frequent interindividual genetic differences in the human genome. The frequency and distribution of de novo somatic structural variants in normal cells is, however, poorly explored. Using age-stratified cohorts of 318 monozygotic (MZ) twins and 296 single-born subjects, we describe age-related accumulation of copy-number variation in the nuclear genomes in vivo and frequency changes for both megabase- and kilobase-range variants. Megabase-range aberrations were found in 3.4% (9 of 264) of subjects ≥60 years old; these subjects included 78 MZ twin pairs and 108 single-born individuals. No such findings were observed in 81 MZ pairs or 180 single-born subjects who were ≤55 years old. Recurrent region- and gene-specific mutations, mostly deletions, were observed. Longitudinal analyses of 43 subjects whose data were collected 7-19 years apart suggest considerable variation in the rate of accumulation of clones carrying structural changes. Furthermore, the longitudinal analysis of individuals with structural aberrations suggests that there is a natural self-removal of aberrant cell clones from peripheral blood. In three healthy subjects, we detected somatic aberrations characteristic of patients with myelodysplastic syndrome. The recurrent rearrangements uncovered here are candidates for common age-related defects in human blood cells. We anticipate that extension of these results will allow determination of the genetic age of different somatic-cell lineages and estimation of possible individual differences between genetic and chronological age. Our work might also help to explain the cause of an age-related reduction in the number of cell clones in the blood; such a reduction is one of the hallmarks of immunosenescence.
Copyright © 2012 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures
Figure 1
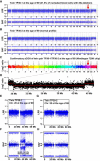
Two Examples of Megabase-Range De Novo Somatic Aberrations (A) A normal profile of MZ twin TP25-1. (B) A 32.5 Mb deletion on 5q is shown in nucleated blood cells of co-twin TP25-2. This deletion was uncovered with LRR data from the Illumina SNP array. (C and D) The BAF profiles of twins TP25-1 (C) and TP25-2 (D). The qPCR experiments showed that 66.2% of nucleated blood cells in TP25-2 had the 5q deletion (i.e., 33.1% fewer copies of the DNA segment, Figure 5). The R-package-MAD (Mosaic Alteration Detection) analysis of the Illumina data suggested that 50.5% of the cells had the 5q deletion when the subjects were 77 years old. (E) The deviation of BAF values from 0.5 (the allelic fraction of intensity at each heterozygous SNP) was plotted, and the percentage of cells with the 5q deletion was higher when the subjects were 77 years old than when they were 70 years old (t test: p < 0.001). This slow increase in aberrant clones was also supported by the MAD estimate of 48.3% of cells detected when the subjects were 70 years old. The size and position of this deletion is typical of patients with myelodysplastic syndrome (MDS). (F) A confirmatory array-CGH experiment. (G–K) Another large somatic event: a terminal CNNLOH encompassing 103 Mb of 4q in ULSAM-697. The LRR and BAF data from Illumina SNP genotyping of samples collected when the subjects were 71, 82, 88, and 90 years old are plotted in (G), (H), (I), and (J), respectively. Percentages of cells with the aberration were calculated with the MAD package and are given for each panel. (K) The proportion of cells with the 4q aberration changes with time, and the changes are significantly different between all samplings at different ages (ANOVA: F(3,25935) = 39087, p < 0.001; Tukey's test for multiple comparisons). Figure S8 shows other analysis details of the samples collected from ULSAM-697 when he was 90 years old. These analyses include those of fibroblasts and three types of sorted blood cells. The analysis of samples obtained when the subjects were 90 years old was performed in duplicate experiments on Illumina 1M-Duo and Omni-Express arrays.
Figure 2
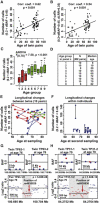
Age-Related Accumulation of Small Somatic Structural Rearrangements in 87 Pairs of MZ Twins (A and B) Linear regression analyses showing that the number of calls increases with age in MZ twin pairs when ΔBAF values are between 0.2 and 0.45 as well as when ΔBAF values are between 0.2 and 0.45 and when the LRR deviation is >0.35. Each dot represents data from one MZ twin pair. Details regarding the filtering algorithms used are shown in Figure S1. (C and D) An analysis of statistical significance for nine age groups of MZ twin pairs when ΔBAF values are between 0.2 and 0.45. (E and F) Longitudinal data analyses comparing the number of ΔBAF reports (between 0.2 and 0.45) of 18 twin pairs that were sampled twice, 10 years apart. Each point in the plot represents the number of differences within one MZ pair (E). Each line (plotted between the two time points for the same MZ pair) thus represents the change over time of the number of differences within a pair (blue line, increase; red line, decrease; green line, no change). The intraindividual changes for each twin over a period of 10 years are shown in (F). The x axis shows individual ages at the later sampling. On the y axis, the number of differences found between the two samples from the same person at the two time points is shown, and vertical lines connect co-twins. (G and H) Validation of copy-number imbalance between MZ twins in two pairs (chromosomes 10 and 6, respectively), which were detected by the ΔBAF analysis. The small boxes at the top of both (G) and (H) display original data from Illumina arrays for pairs TP63-1/TP63-2 and TP31-1/TP32-2, respectively. The larger boxes at the bottom of (G) and (H) display raw data from Nimblegen tiling-path 135K array for these two twin pairs. Each line is drawn to scale and represents data from one oligonucleotide probe. Statistical significance for the results of the Nimblegen array was calculated with the Mann-Whitney U test; values were analyzed for the region of interest (shaded) and for both areas on either side of the control regions. Twenty additional examples of validation experiments are shown in Figure S6. There was no difference between the rates of validation success for the young (n = 8) and old (n = 26) MZ pairs used in these experiments (t test: t = 0.7062, p value = 0.4819), supporting the results from linear-regression analyses. The detailed description of the Nimblegen array is provided in Figure S6 and Table S4.
Figure 3
An Example of a Somatic Megabase-Range Aberration (A, E, and F) A deletion encompassing 12.9 Mb of 20q in MZ twin TP30-1 was sampled when she was 69 years old. (B, G, and H) The normal profile of co-twin TP30-2, as detected by LRR and BAF after Illumina SNP array genotyping. R-package-MAD analysis of the Illumina data suggested that 41.5% of the blood cells had the 20q deletion. qPCR validation experiments confirmed this result by showing 39.6% aberrant cells (i.e., 19.8% fewer copies of the DNA segment, Figure 5). (C and D) Array-CGH validation experiments also confirmed the copy-number variation. The genetic change in MZ twin TP30-1 is another example of an MDS-like aberration, which was uncovered in a subject without a clinical diagnosis of MDS.
Figure 4
Longitudinal Analysis of ULSAM-340, a Single-Born Subject Containing a 13.8 Mb Deletion on 20q, as Detected by LRR and BAF with the Illumina SNP Array The size and position of this deletion is typical of MDS patients. This subject, however, has not been diagnosed with MDS. When the patient was 71 years old, the deletion was only carried by a small proportion of blood cells and was barely detectable, and neither Nexus Copy Number software nor R-package MAD reported this aberration at this age (A, D, and E). R-package MAD suggested that 50.7% of the nucleated cells had the deletion when ULSAM-340 was 75 years old (B, F, and G) and that when he was 88 years old, the corresponding proportion of cells was 36.1% (C, H, and I). qPCR validation experiments showed that the sample taken when the patient was 88 years old contained 14.5% fewer copies of DNA in the segment as compared to the sample taken when he was 75 years old (Figure 5). The deviations from 0.5 of the BAF values within the deleted region in the three different sampling stages are illustrated in (J).
Figure 5
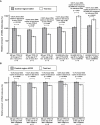
Validation of de novo CNVs by qPCR with SYBR Green Eleven independent qPCR experiments, each composed of multiple (5–11) independent measurements, are shown. The relative number of DNA copies in both test loci (white bars) and the control region UCE3 (gray bars) were plotted. Before we plotted and performed statistical analyses with t tests, we normalized all Ct values by using the control region UCE6. Figure S7 shows the determination of primer efficiency for each of the primer pairs. (A and B) Validations for five large-scale (A) and five small-scale (B) aberrations. The dotted line drawn at 100% represents the copy-number state in control DNA (i.e., that from the normal MZ co-twin, or human female control DNA, or DNA from the same subject sampled at another age), and error bars indicate standard error of means. (A) The 5q deletion in twin TP25-2 (Figure 1) was validated with two primer pairs (41.1 and 42.1) designed within the deleted region. In total, ten independent qPCR experiments showed that ∼66.2% of all nucleated blood cells in TP25-2 had the 5q deletion (i.e., an average of 33.1% [30.8% with primer pair 41.1 and 35.4% with primer pair 42.1] fewer copies of the DNA segment). Similarly, the 20q deletion in twin TP30-1 (Figure 3) was validated with primer pair 45.1 in five experiments. The 19.8% fewer DNA copies found in the test locus indicates that 39.6% of the nucleated blood cells had the deletion. For ULSAM-340, the array data indicated a longitudinal somatic change in the number of cells carrying the 20q deletion. Six independent qPCR experiments comparing DNA sampled when ULSAM-340 was 75 and 88 years old showed that the subject had 14.5% fewer copies of the DNA segment when he was 88 years old. In ULSAM-102, the Illumina array identified a duplication event on both chromosomes 1 and 8 (Figure S9). Given that the proportion of cells with a gained segment in this subject was relatively stable over time, we used human female genomic DNA as control DNA in these experiments. The qPCR experiments validated both somatic CNVs. (B) qPCR validation of five loci with small-scale de novo CNVs within MZ twins. These loci were identified by Illumina array genotyping and were confirmed on the Nimblegen 135K array (see also Figures 2G, 2H, and S6). The layout of this panel is similar to that of (A), described above. For example, the first locus (rs6928830) illustrates de novo CNVs in twin TP31-1 (Figure 2H).
Similar articles
- CNV Concordance in 1,097 MZ Twin Pairs.
Abdellaoui A, Ehli EA, Hottenga JJ, Weber Z, Mbarek H, Willemsen G, van Beijsterveldt T, Brooks A, Hudziak JJ, Sullivan PF, de Geus EJ, Davies GE, Boomsma DI. Abdellaoui A, et al. Twin Res Hum Genet. 2015 Feb;18(1):1-12. doi: 10.1017/thg.2014.86. Epub 2015 Jan 12. Twin Res Hum Genet. 2015. PMID: 25578775 - Aging as accelerated accumulation of somatic variants: whole-genome sequencing of centenarian and middle-aged monozygotic twin pairs.
Ye K, Beekman M, Lameijer EW, Zhang Y, Moed MH, van den Akker EB, Deelen J, Houwing-Duistermaat JJ, Kremer D, Anvar SY, Laros JF, Jones D, Raine K, Blackburne B, Potluri S, Long Q, Guryev V, van der Breggen R, Westendorp RG, 't Hoen PA, den Dunnen J, van Ommen GJ, Willemsen G, Pitts SJ, Cox DR, Ning Z, Boomsma DI, Slagboom PE. Ye K, et al. Twin Res Hum Genet. 2013 Dec;16(6):1026-32. doi: 10.1017/thg.2013.73. Epub 2013 Nov 4. Twin Res Hum Genet. 2013. PMID: 24182360 - Ontogenetic de novo copy number variations (CNVs) as a source of genetic individuality: studies on two families with MZD twins for schizophrenia.
Maiti S, Kumar KH, Castellani CA, O'Reilly R, Singh SM. Maiti S, et al. PLoS One. 2011 Mar 2;6(3):e17125. doi: 10.1371/journal.pone.0017125. PLoS One. 2011. PMID: 21399695 Free PMC article. - Structural genetic variation in the context of somatic mosaicism.
Dumanski JP, Piotrowski A. Dumanski JP, et al. Methods Mol Biol. 2012;838:249-72. doi: 10.1007/978-1-61779-507-7_12. Methods Mol Biol. 2012. PMID: 22228016 Review. - Detection of copy number alterations in acute myeloid leukemia and myelodysplastic syndromes.
Jacoby MA, Walter MJ. Jacoby MA, et al. Expert Rev Mol Diagn. 2012 Apr;12(3):253-64. doi: 10.1586/erm.12.18. Expert Rev Mol Diagn. 2012. PMID: 22468816 Review.
Cited by
- Deletions Rate-Limit Breast and Ovarian Cancer Initiation.
Houlahan KE, Bihie M, Contreras JG, Fulop DJ, Lopez G, Huang HH, Van Loo P, Curtis C, Boutros PC, Huang KL. Houlahan KE, et al. bioRxiv [Preprint]. 2024 Oct 21:2024.10.17.618945. doi: 10.1101/2024.10.17.618945. bioRxiv. 2024. PMID: 39484366 Free PMC article. Preprint. - Association of autosomal mosaic chromosomal alterations with risk of bladder cancer in Chinese adults: a prospective cohort study.
Song M, Han Y, Zhao Y, Lv J, Yu C, Pei P, Yang L, Millwood IY, Walters RG, Chen Y, Du H, Yang X, Yao W, Chen J, Chen Z, Genovese G, Terao C, Li L, Sun D; China Kadoorie Biobank Collaborative Group. Song M, et al. Cell Death Dis. 2024 Sep 30;15(9):706. doi: 10.1038/s41419-024-07087-6. Cell Death Dis. 2024. PMID: 39349436 Free PMC article. - Cell-type-specific consequences of mosaic structural variants in hematopoietic stem and progenitor cells.
Grimes K, Jeong H, Amoah A, Xu N, Niemann J, Raeder B, Hasenfeld P, Stober C, Rausch T, Benito E, Jann JC, Nowak D, Emini R, Hoenicka M, Liebold A, Ho A, Shuai S, Geiger H, Sanders AD, Korbel JO. Grimes K, et al. Nat Genet. 2024 Jun;56(6):1134-1146. doi: 10.1038/s41588-024-01754-2. Epub 2024 May 28. Nat Genet. 2024. PMID: 38806714 Free PMC article. - Genome integrity as a potential index of longevity in Ashkenazi Centenarian's families.
Andrawus M, David GB, Terziyska I, Sharvit L, Bergman A, Barzilai N, Raj SM, Govindaraju DR, Atzmon G. Andrawus M, et al. Geroscience. 2024 Oct;46(5):4147-4162. doi: 10.1007/s11357-024-01178-0. Epub 2024 May 9. Geroscience. 2024. PMID: 38724875 Free PMC article. - Copy Number Variations in Pancreatic Cancer: From Biological Significance to Clinical Utility.
Oketch DJA, Giulietti M, Piva F. Oketch DJA, et al. Int J Mol Sci. 2023 Dec 27;25(1):391. doi: 10.3390/ijms25010391. Int J Mol Sci. 2023. PMID: 38203561 Free PMC article. Review.
References
- van Ommen G.J. Frequency of new copy number variation in humans. Nat. Genet. 2005;37:333–334. - PubMed
- Lupski J.R. Genomic rearrangements and sporadic disease. Nat. Genet. 2007;39(7 Suppl):S43–S47. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources