Structural basis for recognition of H3K56-acetylated histone H3-H4 by the chaperone Rtt106 - PubMed (original) (raw)
Structural basis for recognition of H3K56-acetylated histone H3-H4 by the chaperone Rtt106
Dan Su et al. Nature. 2012.
Abstract
Dynamic variations in the structure of chromatin influence virtually all DNA-related processes in eukaryotes and are controlled in part by post-translational modifications of histones. One such modification, the acetylation of lysine 56 (H3K56ac) in the amino-terminal α-helix (αN) of histone H3, has been implicated in the regulation of nucleosome assembly during DNA replication and repair, and nucleosome disassembly during gene transcription. In Saccharomyces cerevisiae, the histone chaperone Rtt106 contributes to the deposition of newly synthesized H3K56ac-carrying H3-H4 complex on replicating DNA, but it is unclear how Rtt106 binds H3-H4 and specifically recognizes H3K56ac as there is no apparent acetylated lysine reader domain in Rtt106. Here, we show that two domains of Rtt106 are involved in a combinatorial recognition of H3-H4. An N-terminal domain homodimerizes and interacts with H3-H4 independently of acetylation while a double pleckstrin-homology (PH) domain binds the K56-containing region of H3. Affinity is markedly enhanced upon acetylation of K56, an effect that is probably due to increased conformational entropy of the αN helix of H3. Our data support a mode of interaction where the N-terminal homodimeric domain of Rtt106 intercalates between the two H3-H4 components of the (H3-H4)(2) tetramer while two double PH domains in the Rtt106 dimer interact with each of the two H3K56ac sites in (H3-H4)(2). We show that the Rtt106-(H3-H4)(2) interaction is important for gene silencing and the DNA damage response.
Figures
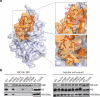
Figure 1. 3D structures of Rtt106 dimeric and double PH domains and their interaction with histones
a, NMR structure of Rtt106 dimeric region (residues 1–67) with the hydrophobic residues constituting the dimerization interface in stick representation. Two protomers are in blue and green. Residues 43–67 are disordered and omitted for clarity. b, Crystal structure of Rtt106 double PH domain (Rtt106PH, residues 68–301) with H3K56ac peptide-binding surface in flesh. c, ITC results (top, raw titration data; bottom, integrated heat measurements ) for the interaction of Rtt106DD–Rtt106PH (residues 1–301) with non-acetylated (H3–H4)2 (black) and K56-acetylated (H3–H4)2 (red). For the former interaction, a two-site binding model (dissociation constants _K_d1 and _K_d2) was used. For the latter, an activation step accounting for the effect of acetylation was incorporated in the two-site binding model (_K_d1ac and _K_d2ac). _K_ds are reported with s.d. determined by nonlinear least-squares analysis. The light blue envelop represents simulated data for _K_d2ac varying from 0.01 to 0.1 μM and _K_d1ac = 0.4 μM. d, ITC data for the interaction of Rtt106DD (residues 1–42) with (H3–H4)2. Stoichiometry n and _K_d are indicated. Data for two mutant forms of Rtt106DD, D7K, E11K and E29K, E32K, E33K, are also shown. e, 1H–15N HSQC spectra of H3K56ac peptide-bound (red) versus free (black) Rtt106PH. Perturbed signals are labelled on the spectra.
Figure 2. Identification of a K56ac-binding cleft in Rtt106 and model of Rtt106 in complex with K56-acetylated (H3–H4)2
a, Binding cleft and the side chain of K301 (red) in Rtt106PH and Rtt106PHL. b, Chemical shift changes in Rtt106PH 1H–15N HSQC spectra upon titration with the H3K56ac peptide (from red to purple signals) are compared to the chemical shifts of free Rtt106PHL (black signals) with respect to seven residues in the vicinity of the binding cleft. A214, E215, K216 and I217 belong to the disordered loop adjacent to the binding cleft. c, Structural model of Rtt106 (residues 1–301) in complex with the (H3–H4)2 tetramer. Atomic coordinates of (H3–H4)2 are from the structure of budding yeast nucleosome core particle (PDB access code 1ID3).
Figure 3. Effects of Rtt106PH mutations on H3K56ac interaction
a, Surface representation of Rtt106PH with the H3K56ac peptide-binding region in orange. The upper box represents a mainly hydrophobic area of the binding site whereas the lower box highlights the K56/K56ac binding cleft. Affinities of Rtt106PH mutants for the H3K56ac peptide were measured and _K_ds are reported in Supplementary Table 3. Mutated residues that totally abolish, decrease, have no effect or enhance binding are labelled in red, blue, black and white, respectively. b, Wild-type (WT) and mutant tandem affinity purification (TAP)-tagged full-length Rtt106 were purified from yeast cells and analysed by western blot using indicated antibodies. CBP, calmodulin-binding peptide tag. Rtt106 mutated outside the binding site (T232A) was used as control.
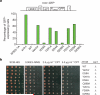
Figure 4. Effects of Rtt106 mutations on HMR silencing and genome stability
a, Schematic of the GFP-based gene silencing reporter assay. The GFP gene (PURA3-GFP) at the silent mating type locus HMR (hmr::GFP) is controlled by the URA3 gene promoter, silencers E and I, and a2 gene. Gene silencing is reported as percentage of yeast cells expressing GFP. One representative of three independent experiments is shown. Silencing is observed in W303-1A control cells, but not in cells lacking the silent information regulator gene SIR3. Expression of wild-type (WT) Rtt106 in cac1Δrtt106Δ cells restores silencing. Expression of Rtt106 mutated in the H3K56ac-binding surface does not or only partially restores silencing. b, Mutations in the H3 binding sites of Rtt106 enhance the DNA-damage sensitivity of cac1Δ mutant cells. Cells of the indicated genotypes were spotted onto media lacking histidine (SCM-HIS) for plasmid selection, without or with MMS or CPT for DNA damage assessment.
References
- Luger K. Dynamic nucleosomes. Chromosome Res. 2006;14:5–16. - PubMed
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
- Corpet A, Almouzni G. Making copies of chromatin: the challenge of nucleosomal organization and epigenetic information. Trends Cell Biol. 2009;19:29–41. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases