Functional and structural characterization of the mammalian TREX-2 complex that links transcription with nuclear messenger RNA export - PubMed (original) (raw)
Functional and structural characterization of the mammalian TREX-2 complex that links transcription with nuclear messenger RNA export
Divyang Jani et al. Nucleic Acids Res. 2012 May.
Abstract
Export of messenger RNA (mRNA) from the nucleus to the cytoplasm is a critical step in the gene expression pathway of eukaryotic cells. Here, we report the functional and structural characterization of the mammalian TREX-2 complex and show how it links transcription/processing with nuclear mRNA export. Mammalian TREX-2 is based on a germinal-centre associated nuclear protein (GANP) scaffold to which ENY2, PCID2 and centrins bind and depletion of any of these components inhibits mRNA export. The crystal structure of the GANP:ENY2 complex shows that two ENY2 chains interact directly with GANP, but they have different orientations from those observed on yeast Sac3. GANP is required to recruit ENY2 to nuclear pore complexes (NPCs), but ENY2 is not necessary to recruit GANP, which requires both its CID and MCM3AP domains, together with nucleoporin Nup153. GANP and ENY2 associate with RNA polymerase II and inhibition of mRNA processing redistributes GANP from NPCs into nuclear foci indicating that mammalian TREX-2 is associated with transcription. Thus, we implicate TREX-2 as an integral component of the mammalian mRNA export machinery where it links transcription and nuclear export by facilitating the transfer of mature mRNPs from the nuclear interior to NPCs.
Figures
Figure 1.
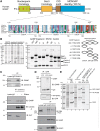
ENY2 and centrin-2 or centrin-3 interact with GANP in vitro and in vivo and PCID2 and DSS1 interact with GANP in vitro. (A) Sequence alignment of the Sac3 CID region showing the conservation of binding sites for Sus1A, Sus1B and Cdc31 (yeast) in orthologues from other species, including human GANP. The numbers below the alignment delimit boundaries for binding sites in yeast and those above are for GANP. (B) Clarified lysates from bacterial cells expressing untagged ENY2, centrin 2 or centrin 3 are shown with the over-expressed protein marked with a black circle. U, un-induced; I, induced. The expression vector for ENY2 shows leaky expression, and so over-expressed protein is evident in the un-induced sample. GST and GST-GANP fragments (containing predicted ENY2 and centrin-binding sites) were co-expressed with either ENY2 (in the case of fragments containing only putative ENY2-binding sites) or centrin (in the case of fragments containing the putative centrin-binding site) and their clarified lysates mixed with clarified lysate-containing centrin or ENY2, respectively. Centrin 3 was used for binding in lanes 1–7 and centrin 2 in lane 8. GST fusion protein-containing complexes were immobilized on glutathione sepharose, washed and analysed by SDS–PAGE and Coomassie staining. Impurities are marked by asterisks. A schematic representation of the GANP fragments used (black bar) depicting their residue numbers and the partner chains (ovals) they would be predicted to bind is also shown. (C) Endogenous centrin-2 and -3 were immunoprecipitated from nuclear extract of HCT116 cells and blotted for GANP, centrin-2 and centrin-3. Centrin-2 was also immunoprecipitated from RNase-treated nuclear extract and blotted for GANP, centrin-2 and ALY. The RNase treatment was effective as centrin-2 association with ALY was abolished following RNase treatment. (D) YFP or YFP-ENY2 was immunoprecipitated with GFP antibody from nuclear extracts of HCT116 cells expressing YFP or YFP-ENY2 and blotted for GFP and GANP. Endogenous ENY2 interacts with both GANP and C-terminal domain of RNA polymerase II. Endogenous GANP and ENY2 were immunoprecipitated from nuclear extract of HCT116 cells and blotted for GANP, ENY2 and RNA polymerase II. (E) Clarified lysates from bacterial cells expressing His-PCID2:DSS1 complex and GST-GANP 686-1003 were applied to Ni-NTA and glutathione sepharose (GS) resins, respectively, and showed that although protein expression levels were low such that over-expressed bands could not be observed in the lysate (I, induced), the appropriate proteins (black circles) could be enriched on the affinity matrices (lanes 1–4). When clarified lysates containing GST-GANP 686-1003 and His-PCID2:DSS1 were mixed, a ternary complex was formed and could be pulled down on Ni-NTA (lane 5). Importantly, the GST-GANP 686-1003 fusion did not have an affinity for Ni-NTA in the absence of His-PCID2:DSS1 (lane 6). Impurities are marked by asterisks.
Figure 2.
Structure of the GANP:ENY2 complex. (A) Overview of the structure of the GANP:ENY2 complex (GANP residues 1162–1233 in green, ENY2A in purple and ENY2B in orange) and comparison to the yeast Sac3 CID complex (PDB 3FWC, (18), Sac3 in red, Sus1A in dark blue, Sus1B in cyan and Cdc31 in yellow). (B) Ribbon representation of ENY2B (orange) and GANP residues 1201–1233 (green) superimposed to Sus1B (cyan) and Sac3 residues 752–788 (red) [PDB 3FWB (18)]. The Cα backbones of Sac3 and GANP have a root-mean-square deviation of 0.39 Å, whereas the Cα backbones of Sus1B and ENY2B have a root-mean-square deviation of 2.10 Å. (C) The two hydrophobic stripes in the ENY2A and ENY2B-binding sites of the GANP helix. The GANP sequence is shown on a helical net, with residues that make a major contribution to the interface shown in bold. Residues forming putative salt bridges or H bonds are shown in red (for acidic) or blue (for basic). Cysteine hydrogen bonds are shown in green. The approximately four-residue repeat sequence that forms the hydrophobic stripes in GANP is broadly the same found in yeast Sac3.
Figure 3.
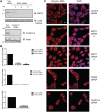
Depletion of TREX-2 components results in nuclear accumulation of poly(A) + RNA. (A) Immunoblotting analysis of HCT116 cells depleted of ENY2 (using two independent siRNAs) or centrin-2. Samples were analysed by Western blotting with the indicated antibodies 72-h post-transfection. (B) Depletion of ENY2, centrin-2 and PCID2 was also confirmed by qRT-PCR using ENY2, centrin-2 and PCID2-specific primers 72-h post-transfection and represent the mean of triplicate readings from three independent depletion experiments ± SD. (C) FISH showing nuclear accumulation of poly(A) + RNA in either ENY2, PCID2 or centrin-2 depleted cells. ENY2-depleted cells were assayed 96-h post-transfection, whereas PCID2 and centrin-2-depleted cells were assayed 72-h post-transfection. Merged image is shown in right panels (Scale bar, 5 µm). Poly(A) + RNA was identified using a Cy3-labelled oligo(dT) probe. Nuclei were stained with DAPI.
Figure 4.
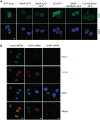
CID and MCM3AP domains of GANP but not ENY2 are required for its NPC association. (A) CID and MCM3AP domains of GANP are required for its NPC association. Various constructs of GANP fused to GFP were transfected into HCT116 cells and localization examined by epifluorescence. GANP-GFP and GANPΔCID-GFP localize to NPCs, but GANPΔMCM3AP-GFP is nuclear. CID + MCM3AP-GFP localizes to NPCs. Nuclei were stained with DAPI (scale bar, 5 µm). (B) GANP is required for ENY2 association with NPCs. In HCT116 cells permeabilized before fixation, GANP and ENY2 co-localize at the NPC. Following depletion of ENY2, GANP NPC staining is retained, but ENY2 staining is abolished. In contrast, GANP depletion results in loss of both GANP and ENY2 staining from NPCs.
Figure 5.
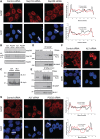
GANP localization is dependent on the early-stage mRNA export factor ALY and Nup153; GANP and ENY2 associate with RNA Pol II. (A) Depletion of Nup153, but not Nup358 redistributes GANP into nuclear foci with less at NPCs. Scanning analysis of GANP intensity in control siRNA treated or Nup153- or Nup358-depleted cells is shown. Nuclei used for scanning and the scanning axis are indicated by white lines. Pairs of nuclei of same scan width as determined by DAPI staining were used for scanning (scale bar, 5 µm). (B) Efficiency of depletion of Nup153 and Nup358 was determined by Western blotting with mAb414 antibody, which recognizes FG-repeat containing nucleoporins. (C) Efficiency of depletion of ALY was monitored by immunoblotting with the indicated antibodies. (D) The interaction between endogenous GANP and the C-terminal domain of RNA polymerase II is increased following RNase treatment. Endogenous GANP was immunoprecipitated from untreated and RNase treated nuclear extract of HCT116 cells and blotted for GANP and RNA polymerase II. (E) Endogenous RNA polymerase II was also immunoprecipitated from untreated and RNase-treated nuclear extract of HCT116 cells and blotted for GANP and RNA polymerase II. (F) Depletion of ALY results in nuclear accumulation of poly(A) + RNA. FISH showing nuclear accumulation of poly(A) + RNA in ALY-depleted cells 72-h post-transfection (scale bar, 5 µm). (G) Depletion of ALY redistributes GANP into nuclear foci with less at NPCs (scale bar, 5 µm). Scanning analysis of GANP intensity in control siRNA treated or ALY- or PCID2-depleted cells is shown and analysed as earlier.
Similar articles
- mRNA export from mammalian cell nuclei is dependent on GANP.
Wickramasinghe VO, McMurtrie PI, Mills AD, Takei Y, Penrhyn-Lowe S, Amagase Y, Main S, Marr J, Stewart M, Laskey RA. Wickramasinghe VO, et al. Curr Biol. 2010 Jan 12;20(1):25-31. doi: 10.1016/j.cub.2009.10.078. Epub 2009 Dec 10. Curr Biol. 2010. PMID: 20005110 Free PMC article. - GANP enhances the efficiency of mRNA nuclear export in mammalian cells.
Wickramasinghe VO, Stewart M, Laskey RA. Wickramasinghe VO, et al. Nucleus. 2010 Sep-Oct;1(5):393-6. doi: 10.4161/nucl.1.5.12351. Nucleus. 2010. PMID: 21326821 Free PMC article. - The human TREX-2 complex is stably associated with the nuclear pore basket.
Umlauf D, Bonnet J, Waharte F, Fournier M, Stierle M, Fischer B, Brino L, Devys D, Tora L. Umlauf D, et al. J Cell Sci. 2013 Jun 15;126(Pt 12):2656-67. doi: 10.1242/jcs.118000. Epub 2013 Apr 16. J Cell Sci. 2013. PMID: 23591820 - Germinal Center B-Cell-Associated Nuclear Protein (GANP) Involved in RNA Metabolism for B Cell Maturation.
Sakaguchi N, Maeda K. Sakaguchi N, et al. Adv Immunol. 2016;131:135-86. doi: 10.1016/bs.ai.2016.02.003. Epub 2016 Mar 29. Adv Immunol. 2016. PMID: 27235683 Review. - The critical role of germinal center-associated nuclear protein in cell biology, immunohematology, and hematolymphoid oncogenesis.
Sakai Y, Phimsen S, Okada S, Kuwahara K. Sakai Y, et al. Exp Hematol. 2020 Oct;90:30-38. doi: 10.1016/j.exphem.2020.08.007. Epub 2020 Aug 20. Exp Hematol. 2020. PMID: 32827560 Review.
Cited by
- PCID2 dysregulates transcription and viral RNA processing to promote HIV-1 latency.
Crespo R, Ne E, Reinders J, Meier JIJ, Li C, Jansen S, Górska A, Koçer S, Kan TW, Doff W, Dekkers D, Demmers J, Palstra RJ, Rao S, Mahmoudi T. Crespo R, et al. iScience. 2024 Feb 6;27(3):109152. doi: 10.1016/j.isci.2024.109152. eCollection 2024 Mar 15. iScience. 2024. PMID: 38384833 Free PMC article. - Control of mammalian gene expression by selective mRNA export.
Wickramasinghe VO, Laskey RA. Wickramasinghe VO, et al. Nat Rev Mol Cell Biol. 2015 Jul;16(7):431-42. doi: 10.1038/nrm4010. Epub 2015 Jun 17. Nat Rev Mol Cell Biol. 2015. PMID: 26081607 Review. - Proteomics Uncovers Novel Components of an Interactive Protein Network Supporting RNA Export in Trypanosomes.
Inoue AH, Domingues PF, Serpeloni M, Hiraiwa PM, Vidal NM, Butterfield ER, Del Pino RC, Ludwig A, Boehm C, Field MC, Ávila AR. Inoue AH, et al. Mol Cell Proteomics. 2022 Mar;21(3):100208. doi: 10.1016/j.mcpro.2022.100208. Epub 2022 Jan 26. Mol Cell Proteomics. 2022. PMID: 35091090 Free PMC article. - Luzp4 defines a new mRNA export pathway in cancer cells.
Viphakone N, Cumberbatch MG, Livingstone MJ, Heath PR, Dickman MJ, Catto JW, Wilson SA. Viphakone N, et al. Nucleic Acids Res. 2015 Feb 27;43(4):2353-66. doi: 10.1093/nar/gkv070. Epub 2015 Feb 6. Nucleic Acids Res. 2015. PMID: 25662211 Free PMC article. - Structural Characterization of the Chaetomium thermophilum TREX-2 Complex and its Interaction with the mRNA Nuclear Export Factor Mex67:Mtr2.
Dimitrova L, Valkov E, Aibara S, Flemming D, McLaughlin SH, Hurt E, Stewart M. Dimitrova L, et al. Structure. 2015 Jul 7;23(7):1246-57. doi: 10.1016/j.str.2015.05.002. Epub 2015 Jun 4. Structure. 2015. PMID: 26051714 Free PMC article.
References
- Tutucci E, Stutz F. Keeping mRNPs in check during assembly and nuclear export. Nat. Rev. Mol. Cell Biol. 2011;12:377–384. - PubMed
- Rodriguez-Navarro S, Hurt E. Linking gene regulation to mRNA production and export. Curr. Opin. Cell Biol. 2011;23:302–309. - PubMed
- Stewart M. Nuclear export of mRNA. Trends Biochem. Sci. 2010;35:609–617. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MC_U105178939/MRC_/Medical Research Council/United Kingdom
- U105178939/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous