Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane binding protein - PubMed (original) (raw)
doi: 10.1038/cr.2012.24. Epub 2012 Feb 7.
Wooyoung Choi, Wanqiu Hu, Na Mi, Qiang Guo, Meisheng Ma, Mei Liu, Yuan Tian, Peilong Lu, Feng-Liang Wang, Haiteng Deng, Lei Liu, Ning Gao, Li Yu, Yigong Shi
Affiliations
- PMID: 22310240
- PMCID: PMC3292424
- DOI: 10.1038/cr.2012.24
Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane binding protein
Weijiao Huang et al. Cell Res. 2012 Mar.
Abstract
The Beclin 1 gene is a haplo-insufficient tumor suppressor and plays an essential role in autophagy. However, the molecular mechanism by which Beclin 1 functions remains largely unknown. Here we report the crystal structure of the evolutionarily conserved domain (ECD) of Beclin 1 at 1.6 Å resolution. Beclin 1 ECD exhibits a previously unreported fold, with three structural repeats arranged symmetrically around a central axis. Beclin 1 ECD defines a novel class of membrane-binding domain, with a strong preference for lipid membrane enriched with cardiolipin. The tip of a surface loop in Beclin 1 ECD, comprising three aromatic amino acids, acts as a hydrophobic finger to associate with lipid membrane, consequently resulting in the deformation of membrane and liposomes. Mutation of these aromatic residues rendered Beclin 1 unable to stably associate with lipid membrane in vitro and unable to fully rescue autophagy in Beclin 1-knockdown cells in vivo. These observations form an important framework for deciphering the biological functions of Beclin 1.
Figures
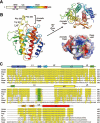
Figure 1
Structure of the Beclin 1 ECD. (A) Beclin 1 contains a BH3 domain, a coiled coil and an ECD. Rainbow color for the ECD domain is preserved in panel B. (B) Overall structure of the ECD (residues 248-447) of human Beclin 1. The structure is shown in rainbow color, with the N- and C-termini colored blue and red, respectively. An aromatic finger, comprising Phe359, Phe360 and Trp361, protrudes from the core structure. Beclin 1 ECD contains three structural repeats, each comprising a pair of β-strands and a long α-helix (top right). A deep pocket is located next to the aromatic finger (bottom right). (C) Sequence alignment of Beclin 1 homologs from multiple organisms. Conserved sequences are colored yellow. The three hydrophobic residues in the aromatic finger, highlighted in green, are highly conserved in all organisms except yeast. Secondary structural elements are indicated above the sequences. All structural images were generated using PyMol .
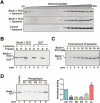
Figure 2
Beclin 1 ECD directly binds to lipid membrane. (A) Beclin 1 ECD associates with liposome by iodixanol gradient sedimentation analysis. Liposome co-sediments with low percentage of iodixanol, where Beclin 1 ECD was also detected (top panel). Free Beclin 1 ECD co-sediments with high percentage of iodixanol (top and middle panels). The control protein Nampt failed to co-sediment with liposome (bottom panel). (B) Beclin 1 ECD binds to liposome by ultracentrifugation analysis. Beclin 1 ECD or GST was ultracentrifuged in the presence or absence of liposome. The pellet and supernatant fractions were analyzed by SDS-PAGE and coomassie staining. (C) Analysis of liposome binding by Beclin 1 ECD. A fixed amount of liposome was ultracentrifuged in the presence of increasing amounts of Beclin 1 ECD. The pellets were analyzed by SDS-PAGE and coomassie staining. (D) Beclin 1 ECD preferentially binds to liposome enriched with cardiolipin. Liposomes of five distinct compositions were prepared, each containing fixed components (70% PC and 20% PE) and a variable component (10% each of the five specific phospholipids). Binding to Beclin 1 ECD was analyzed for these five liposomes. The control liposome has the same phospholipid composition as that of the Xenopus mitochondria .
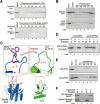
Figure 3
Mutations in the aromatic finger of Beclin 1 ECD cripple its binding to mitochondria and liposomes. (A) The WT Beclin 1 ECD, but not the finger mutant (F359D/F360D/W361D), bound to liposome by ultracentrifugation analysis. (B) The WT Beclin 1 ECD, but not the finger mutant (F359D/F360D/W361D), exhibited a strong preference for cardiolipin-enriched liposomes. (C) A close-up view of the rationale for engineering a LC3 variant with grafted aromatic finger from Beclin 1. Amino acids 72-78 of LC3 constitute a surface loop with features similar to those of residues 354-363 of Beclin 1 ECD. Notably, the main chain Cα-Cα distances are similar in both cases. (D) The LC3 variant with engrafted aromatic finger, but not the WT LC3, associated with liposomes. S: supernatant; P: pellet (i.e., liposome fraction). (E) The LC3 variant with engrafted aromatic finger, but not the WT LC3, exhibited a strong preference for cardiolipin-enriched liposomes. (F) The full-length WT Beclin 1, but not the finger mutant (F359D/F360D/W361D), bound to liposome. S: supernatant; P: pellet (i.e., liposome fraction).
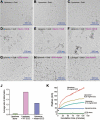
Figure 4
The Beclin 1 ECD deforms liposome. Free liposomes (panels A-C), liposomes pre-incubated with WT ECD (panels D-F), and liposomes pre-incubated with mutant ECD (panels G-I) were treated with nanogold particles and visualized under cryo-electron microscope. Beclin 1 ECD contains a 6×His tag, and the nanogold particle is linked to Ni2+-NTA. The WT Beclin 1 ECD, but not the mutant ECD, allowed the nanogold particles to be concentrated in the areas of liposome deformation, which might be liposome fusion, vesicle budding, and/or membrane vesiculation. (J) Quantitative analysis of liposome deformation by Beclin 1 ECD. The extent of liposome deformation is defined by the ratio of the longest dimension over the shortest dimension for each liposome. These ratios for 65 randomly chosen liposomes for each of three categories, control (no ECD), with WT ECD, and with mutant ECD, were measured and averaged. Liposomes incubated with WT ECD exhibited a larger average ratio than that for liposomes incubated with mutant ECD. **(K)**Incubation with Beclin 1 ECD led to increased radius for the liposomes. The increase is correlated with the concentrations of Beclin 1 ECD.
Figure 5
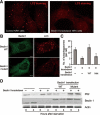
The aromatic finger of Beclin 1 plays an important role in autophagy.(A) Beclin 1-knockdown cells exhibited a markedly reduced level of autophagy upon starvation. Control and Beclin 1 stable knockdown NRK cells were stained with anti-LC3 antibody 4 h after starvation. Scale bar, 5 μm. (B) The WT Beclin 1, but not the finger mutant (F359D/F360D/W361D), rescued the autophagy defect in NRK cells. Beclin 1 stable knockdown NRK cells were transfected with WT or mutant Beclin 1-YFP. 24 h after transfection, cells were starved for 4 h. Then the cells were stained with anti-GFP or anti-LC3 antibody. Scale bar, 5 μm.(C) Average numbers of autophagosome per cell are shown for panels A and B. 50 cells were counted for each sample.(D) NRK cells transfected with the WT Beclin 1, but not with the finger mutant (F359D/F360D/W361D), had significant autophagic degradation of p62, a hallmark of autophagy. Cells from panels A and B were analyzed by western blot using antibodies specific for p62, Beclin 1 and actin. The position of transfected Beclin 1 is higher because Beclin 1 was fused to YFP.
Figure 6
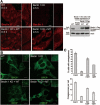
The aromatic finger of Beclin 1 might be involved in omegasome formation.(A) The WT, full-length Beclin 1 exhibited punctate staining 30 min after starvation. Shown here are staining pattern for Beclin 1 in normal HeLa cells (WT), Beclin 1 stable knockdown HeLa cells (Beclin 1 KD), WT Beclin 1 stably expressing Beclin 1 KD cells (Beclin 1 KD+WT), mutant Beclin 1 stably expressing Beclin 1 KD cells (Beclin 1KD+MT). Scale bar, 10 μm. The right panel shows the expression levels of WT and mutant Beclin 1 in the 4 different types of cells. (B) The cellular localization of DFCP was restored by WT Beclin 1, but not mutant. Shown here are cellular staining pattern for DFCP in normal HeLa cells (WT), Beclin 1 KD cells, Beclin 1 KD+WT cells transfected with DFCP-GFP, and Beclin 1 KD+MT cells transfected with DFCP-GFP. 18 h after DFCP transfection, cells were starved for 0.5 h and stained for antibody against GFP. Scale bar, 5 μm. (C) The WT Beclin 1, but not the mutant, supported formation of a greater number of omegasomes (upper panel). Importantly, for the omegasome-containing cells, the average number of omegasomes in Beclin 1 KD+WT cells is ∼11.3, 57% more than that (7.2) in Beclin 1 KD+MT cells. Cells from panel B were quantified. 50 cells were counted for each category. Error bars represent standard deviation. For the top panel, the statistical P values are 0.0001/0.0055/0.0001 between “Cont” and “KD”/“KD+WT”/“KD+MT”, 0.0001/0.0001 between “KD” and “KD+WT”/“KD+MT”, and 0.0015 between “KD+WT” and “KD+MT”. For the bottom panel, the statistical P values are 0.0001/0.0022/0.0012 between “Cont” and “KD”/“KD+WT”/“KD+MT”, 0.0001/0.0216 between “KD” and “KD+WT”/“KD+MT”, and 0.0163 between “KD+WT” and “KD+MT”.
Similar articles
- Beclin 1 cleavage by caspase-3 inactivates autophagy and promotes apoptosis.
Zhu Y, Zhao L, Liu L, Gao P, Tian W, Wang X, Jin H, Xu H, Chen Q. Zhu Y, et al. Protein Cell. 2010 May;1(5):468-77. doi: 10.1007/s13238-010-0048-4. Epub 2010 Jun 4. Protein Cell. 2010. PMID: 21203962 Free PMC article. - DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy.
Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R, Kimchi A. Zalckvar E, et al. EMBO Rep. 2009 Mar;10(3):285-92. doi: 10.1038/embor.2008.246. Epub 2009 Jan 30. EMBO Rep. 2009. PMID: 19180116 Free PMC article. - Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1.
Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, Tasdemir E, Pierron G, Troulinaki K, Tavernarakis N, Hickman JA, Geneste O, Kroemer G. Maiuri MC, et al. EMBO J. 2007 May 16;26(10):2527-39. doi: 10.1038/sj.emboj.7601689. Epub 2007 Apr 19. EMBO J. 2007. PMID: 17446862 Free PMC article. - Impaired autophagy and APP processing in Alzheimer's disease: The potential role of Beclin 1 interactome.
Salminen A, Kaarniranta K, Kauppinen A, Ojala J, Haapasalo A, Soininen H, Hiltunen M. Salminen A, et al. Prog Neurobiol. 2013 Jul-Aug;106-107:33-54. doi: 10.1016/j.pneurobio.2013.06.002. Epub 2013 Jul 1. Prog Neurobiol. 2013. PMID: 23827971 Review. - Beclin 1 biology and its role in heart disease.
Zhu H, He L. Zhu H, et al. Curr Cardiol Rev. 2015;11(3):229-37. doi: 10.2174/1573403x10666141106104606. Curr Cardiol Rev. 2015. PMID: 25373623 Free PMC article. Review.
Cited by
- Human Atg8-cardiolipin interactions in mitophagy: Specific properties of LC3B, GABARAPL2 and GABARAP.
Antón Z, Landajuela A, Hervás JH, Montes LR, Hernández-Tiedra S, Velasco G, Goñi FM, Alonso A. Antón Z, et al. Autophagy. 2016 Dec;12(12):2386-2403. doi: 10.1080/15548627.2016.1240856. Epub 2016 Oct 20. Autophagy. 2016. PMID: 27764541 Free PMC article. - Role of the inositol 1,4,5-trisphosphate receptor/Ca2+-release channel in autophagy.
Parys JB, Decuypere JP, Bultynck G. Parys JB, et al. Cell Commun Signal. 2012 Jul 6;10(1):17. doi: 10.1186/1478-811X-10-17. Cell Commun Signal. 2012. PMID: 22770472 Free PMC article. - Self-eating with your fingers.
Klionsky DJ, Hurley JH. Klionsky DJ, et al. Cell Res. 2012 May;22(5):783-5. doi: 10.1038/cr.2012.39. Epub 2012 Mar 13. Cell Res. 2012. PMID: 22410794 Free PMC article. No abstract available. - Structural insights into BCL2 pro-survival protein interactions with the key autophagy regulator BECN1 following phosphorylation by STK4/MST1.
Lee EF, Smith NA, Soares da Costa TP, Meftahi N, Yao S, Harris TJ, Tran S, Pettikiriarachchi A, Perugini MA, Keizer DW, Evangelista M, Smith BJ, Fairlie WD. Lee EF, et al. Autophagy. 2019 May;15(5):785-795. doi: 10.1080/15548627.2018.1564557. Epub 2019 Jan 9. Autophagy. 2019. PMID: 30626284 Free PMC article. - Effect of autophagy-associated proteins on the arecoline-induced liver injury in mice.
Wang X, Song X, Si Y, Xia J, Wang B, Wang P. Wang X, et al. Exp Ther Med. 2018 Oct;16(4):3041-3049. doi: 10.3892/etm.2018.6564. Epub 2018 Aug 2. Exp Ther Med. 2018. PMID: 30214523 Free PMC article.
References
- Aita VM, Liang XH, Murty VV, et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. - PubMed
- Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources