DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes - PubMed (original) (raw)
. 2012 Feb 7;484(7392):69-74.
doi: 10.1038/nature10909.
Wolfgang Resch, Arito Yamane, Isaac Klein, Kyong-Rim Kieffer-Kwon, Mila Jankovic, Thiago Oliveira, Anne Bothmer, Ty C Voss, Camilo Ansarah-Sobrinho, Ewy Mathe, Genqing Liang, Jesse Cobell, Hirotaka Nakahashi, Davide F Robbiani, Andre Nussenzweig, Gordon L Hager, Michel C Nussenzweig, Rafael Casellas
Affiliations
- PMID: 22314321
- PMCID: PMC3459314
- DOI: 10.1038/nature10909
DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes
Ofir Hakim et al. Nature. 2012.
Abstract
Recurrent chromosomal translocations underlie both haematopoietic and solid tumours. Their origin has been ascribed to selection of random rearrangements, targeted DNA damage, or frequent nuclear interactions between translocation partners; however, the relative contribution of each of these elements has not been measured directly or on a large scale. Here we examine the role of nuclear architecture and frequency of DNA damage in the genesis of chromosomal translocations by measuring these parameters simultaneously in cultured mouse B lymphocytes. In the absence of recurrent DNA damage, translocations between Igh or Myc and all other genes are directly related to their contact frequency. Conversely, translocations associated with recurrent site-directed DNA damage are proportional to the rate of DNA break formation, as measured by replication protein A accumulation at the site of damage. Thus, non-targeted rearrangements reflect nuclear organization whereas DNA break formation governs the location and frequency of recurrent translocations, including those driving B-cell malignancies.
Figures
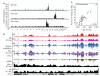
Figure 1. Characterization of the Igh, Myc and Mycn interactomes in B lymphocytes
a, Genome-wide interaction profile of Igh 3′Eα, Myc and Mycn in activated B cells. Plots show the percentage of HindIII fragments carrying 4C-seq reads. b, Contact frequency of Igh with chromosome 5 in activated B cells (lane 1), anti-HEL homozygous activated B cells (lane 2), or resting B cells (lane 3). Histone acetylation (lane 4), RNA Pol II (lane 6) and mRNA (lane 7) density is also shown. Lanes 5 and 8 show Myc and Mycn contacts, respectively. Lane 9 represents Igh contacts in MEFs. c, Comparison of Pol II and Igh 4C-seq data per chromosome normalized as reads per mappable megabase.
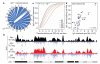
Figure 2. Genomic distribution of AID-independent translocations correlates with nuclear contact profiles
a, Genome-wide view of rearrangements to IghI-SceI in _Aid_−/− B cells. b, Cross-comparison of contacts and translocations between Myc or Igh and mouse chromosome 17 in activated _Aid_−/− B cells. c, Empirical cumulative distribution showing IghI-SceI _Aid_−/− translocations per 200-kb non-overlapping windows as a function of Igh 4C-seq data subdivided as quartiles (Q1–4). NI represents windows with no aligned reads. d, Comparison of AID-independent translocations versus Igh 4C-seq per chromosome per mappable megabase. The degree of correlation is represented by Pearson’s r.
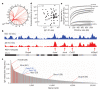
Figure 3. Lack of correlation between translocation hotspots and nuclear architecture
a, Genome-wide hotspots in activated B cells transduced with I-SceI and AID retroviruses (rv). b, Translocation hotspots (bottom lane) and Myc and Igh contacts in chromosome 8. c, Igh contacts with RefSeq genes. Hotspot+ genes are highlighted in red, and for a subset of them the number of translocations is provided in parentheses. The two hotspot− genes (Rplp2 and Rac2) highlighted in blue are discussed in more detailed in panel e. d, Scatter plot showing Igh translocations per hotspot versus contacts. Data are plotted as sequence tags per kb per million sequences (t.p.k.m.). e, Line graph showing the 4C-seq correlation (Spearman’s ρ) between Igh (in various cell types), Rpbp2, Rac2, Myc and Mycn (in activated B cells) versus Igh in activated B cells. The 99% bootstrapping confidence intervals are shown in grey.
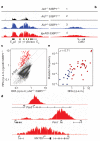
Figure 4. Genome-wide map of AID-mediated DNA damage
a, RPA occupancy at Igh in activated B cells. Genotype and RPA sequence reads per million values are shown. b, Same analysis as in panel a for Cd83. For all tracks, background sequencing was filtered out via a threshold. c, One-hundred and fifty-three RPA islands (red dots) detected in _Igκ_AID _53BP1_−/− B cells fourfold above background (measured in _Aid_−/−53BP1−/− cells). Data are plotted as reads per kb per million sequences (r.p.k.m.). d, RPA islands associated with TSSs from Pax5, Pim1 and Mir155. e, Hypermutation frequency relative to RPA recruitment at TSSs (±2 kb) in a subset of RPA+ (red dots) and RPA− (blue dots) genes. Spearman’s ρ is provided.
Figure 5. AID activity predicts the location and frequency of targeted chromosomal translocations
a, Scatter plot showing the correlation between Igh translocations per hotspot and RPA recruitment. b, Same as panel a but Igh contact frequency is used instead of translocations. c, Upper schematic: distribution of RPA islands (red dots) and translocation hotspots (blue dots) in chromosome 7. Middle: Igh 4C-seq profile demarcating the Nsmce1-Il4ra-Il21r loci. Bottom: RPA islands, translocations and contact frequency for each gene.
Similar articles
- Mechanisms promoting translocations in editing and switching peripheral B cells.
Wang JH, Gostissa M, Yan CT, Goff P, Hickernell T, Hansen E, Difilippantonio S, Wesemann DR, Zarrin AA, Rajewsky K, Nussenzweig A, Alt FW. Wang JH, et al. Nature. 2009 Jul 9;460(7252):231-6. doi: 10.1038/nature08159. Nature. 2009. PMID: 19587764 Free PMC article. - Translocation capture sequencing: a method for high throughput mapping of chromosomal rearrangements.
Oliveira TY, Resch W, Jankovic M, Casellas R, Nussenzweig MC, Klein IA. Oliveira TY, et al. J Immunol Methods. 2012 Jan 31;375(1-2):176-81. doi: 10.1016/j.jim.2011.10.007. Epub 2011 Oct 18. J Immunol Methods. 2012. PMID: 22033343 Free PMC article. - Role of genomic instability and p53 in AID-induced c-myc-Igh translocations.
Ramiro AR, Jankovic M, Callen E, Difilippantonio S, Chen HT, McBride KM, Eisenreich TR, Chen J, Dickins RA, Lowe SW, Nussenzweig A, Nussenzweig MC. Ramiro AR, et al. Nature. 2006 Mar 2;440(7080):105-9. doi: 10.1038/nature04495. Epub 2006 Jan 8. Nature. 2006. PMID: 16400328 Free PMC article. - The origin of recurrent translocations in recombining lymphocytes: a balance between break frequency and nuclear proximity.
Rocha PP, Skok JA. Rocha PP, et al. Curr Opin Cell Biol. 2013 Jun;25(3):365-71. doi: 10.1016/j.ceb.2013.02.007. Epub 2013 Mar 13. Curr Opin Cell Biol. 2013. PMID: 23478218 Free PMC article. Review. - AID and Igh switch region-Myc chromosomal translocations.
Unniraman S, Schatz DG. Unniraman S, et al. DNA Repair (Amst). 2006 Sep 8;5(9-10):1259-64. doi: 10.1016/j.dnarep.2006.05.019. Epub 2006 Jun 19. DNA Repair (Amst). 2006. PMID: 16784901 Review.
Cited by
- 3C-based technologies to study the shape of the genome.
de Laat W, Dekker J. de Laat W, et al. Methods. 2012 Nov;58(3):189-91. doi: 10.1016/j.ymeth.2012.11.005. Methods. 2012. PMID: 23199640 Free PMC article. No abstract available. - Large tandem duplications in cancer result from transcription and DNA replication collisions.
Yang Y, Badura ML, O'Leary PC, Delavan HM, Robinson TM, Egusa EA, Zhong X, Swinderman JT, Li H, Zhang M, Kim M, Ashworth A, Feng FY, Chou J, Yang L. Yang Y, et al. medRxiv [Preprint]. 2024 Jan 10:2023.05.17.23290140. doi: 10.1101/2023.05.17.23290140. medRxiv. 2024. PMID: 38260434 Free PMC article. Updated. Preprint. - The AID-induced DNA damage response in chromatin.
Daniel JA, Nussenzweig A. Daniel JA, et al. Mol Cell. 2013 May 9;50(3):309-21. doi: 10.1016/j.molcel.2013.04.017. Mol Cell. 2013. PMID: 23664375 Free PMC article. Review. - The biogenesis of chromosome translocations.
Roukos V, Misteli T. Roukos V, et al. Nat Cell Biol. 2014 Apr;16(4):293-300. doi: 10.1038/ncb2941. Nat Cell Biol. 2014. PMID: 24691255 Free PMC article. Review. - Investigation of the spatial structure and interactions of the genome at sub-kilobase-pair resolution using T2C.
Kolovos P, Brouwer RWW, Kockx CEM, Lesnussa M, Kepper N, Zuin J, Imam AMA, van de Werken HJG, Wendt KS, Knoch TA, van IJcken WFJ, Grosveld F. Kolovos P, et al. Nat Protoc. 2018 Mar;13(3):459-477. doi: 10.1038/nprot.2017.132. Epub 2018 Feb 8. Nat Protoc. 2018. PMID: 29419817
References
- Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nature Rev. Cancer. 2007;7:233–245. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI037526/AI/NIAID NIH HHS/United States
- Z01 AR041148-03/ImNIH/Intramural NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- Z01 AR041148/ImNIH/Intramural NIH HHS/United States
- AI037526/AI/NIAID NIH HHS/United States
- R37 AI037526/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases