Prasinoviruses of the marine green alga Ostreococcus tauri are mainly species specific - PubMed (original) (raw)
Prasinoviruses of the marine green alga Ostreococcus tauri are mainly species specific
Camille Clerissi et al. J Virol. 2012 Apr.
Abstract
Prasinoviruses infecting unicellular green algae in the order Mamiellales (class Mamiellophyceae) are commonly found in coastal marine waters where their host species frequently abound. We tested 40 Ostreococcus tauri viruses on 13 independently isolated wild-type O. tauri strains, 4 wild-type O. lucimarinus strains, 1 Ostreococcus sp. ("Ostreococcus mediterraneus") clade D strain, and 1 representative species of each of two other related species of Mamiellales, Bathycoccus prasinos and Micromonas pusilla. Thirty-four out of 40 viruses infected only O. tauri, 5 could infect one other species of the Ostreococcus genus, and 1 infected two other Ostreococcus spp., but none of them infected the other genera. We observed that the overall susceptibility pattern of Ostreococcus strains to viruses was related to the size of two host chromosomes known to show intraspecific size variations, that genetically related viruses tended to infect the same host strains, and that viruses carrying inteins were strictly strain specific. Comparison of two complete O. tauri virus proteomes revealed at least three predicted proteins to be candidate viral specificity determinants.
Figures
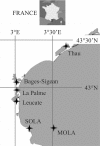
Fig 1
Locations of sampling sites. Four locations are lagoons (Leucate, 42°48′24″N, 03°01′27″E; La Palme, 42°57′18.04″, 3°0′3.56″E; Bages-Sigean, 43°03′14.76″N, 2°59′51.63″E; and Thau, 43°26′01.40″N, 03°39′54.49″E), and the two others are open-sea stations, one coastal (SOLA, 42°29′18″N, 3°8′42″E) and one offshore (MOLA, 42°27′11″N, 3°32′36″E). SOLA is a marine station included in the French marine monitoring network SOMLIT.
Fig 2
Specificity test: lysis spots in a suspension of host cells immobilized in soft agarose. No lysis for C2 and D2; low lysis for A3 and B3; intermediate lysis for A2 and B2; high lysis for A1, B1, C1, D1, C3, and D3.
Fig 3
Specificity patterns. Lines refer to hosts, and columns refer to viruses. −, no lysis; ■, lysis on the host used for the isolation of the virus; ●, high lysis;  , intermediate lysis; ○, low lysis.
, intermediate lysis; ○, low lysis.
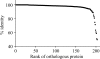
Fig 4
Percent identities between OtV5 and OtV1 orthologous proteins. The most divergent proteins could be involved in the adaptive behavior of O. tauri viruses, particularly in their specificity.
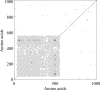
Fig 5
OtV5_074 protein structure. The dot plot shows repetitive motifs for the first 600 amino acids of the OtV5_074 protein sequence. Such motifs have also been found for OtV5_073.
Similar articles
- Marine Prasinoviruses and Their Tiny Plankton Hosts: A Review.
Weynberg KD, Allen MJ, Wilson WH. Weynberg KD, et al. Viruses. 2017 Mar 15;9(3):43. doi: 10.3390/v9030043. Viruses. 2017. PMID: 28294997 Free PMC article. Review. - Isolation of prasinoviruses of the green unicellular algae Ostreococcus spp. on a worldwide geographical scale.
Bellec L, Grimsley N, Desdevises Y. Bellec L, et al. Appl Environ Microbiol. 2010 Jan;76(1):96-101. doi: 10.1128/AEM.01799-09. Epub 2009 Nov 6. Appl Environ Microbiol. 2010. PMID: 19897754 Free PMC article. - Phylogenetic analysis of new Prasinoviruses (Phycodnaviridae) that infect the green unicellular algae Ostreococcus, Bathycoccus and Micromonas.
Bellec L, Grimsley N, Moreau H, Desdevises Y. Bellec L, et al. Environ Microbiol Rep. 2009 Apr;1(2):114-23. doi: 10.1111/j.1758-2229.2009.00015.x. Epub 2009 Feb 19. Environ Microbiol Rep. 2009. PMID: 23765742 - Genome sequence of Ostreococcus tauri virus OtV-2 throws light on the role of picoeukaryote niche separation in the ocean.
Weynberg KD, Allen MJ, Gilg IC, Scanlan DJ, Wilson WH. Weynberg KD, et al. J Virol. 2011 May;85(9):4520-9. doi: 10.1128/JVI.02131-10. Epub 2011 Feb 2. J Virol. 2011. PMID: 21289127 Free PMC article. - The tiny giant of the sea, Ostreococcus's unique adaptations.
Foresi N, De Marco MA, Del Castello F, Ramirez L, Nejamkin A, Calo G, Grimsley N, Correa-Aragunde N, Martínez-Noël GMA. Foresi N, et al. Plant Physiol Biochem. 2024 Jun;211:108661. doi: 10.1016/j.plaphy.2024.108661. Epub 2024 May 1. Plant Physiol Biochem. 2024. PMID: 38735153 Review.
Cited by
- Intermolecular domain swapping induces intein-mediated protein alternative splicing.
Aranko AS, Oeemig JS, Kajander T, Iwaï H. Aranko AS, et al. Nat Chem Biol. 2013 Oct;9(10):616-22. doi: 10.1038/nchembio.1320. Epub 2013 Aug 25. Nat Chem Biol. 2013. PMID: 23974115 - Cophylogenetic interactions between marine viruses and eukaryotic picophytoplankton.
Bellec L, Clerissi C, Edern R, Foulon E, Simon N, Grimsley N, Desdevises Y. Bellec L, et al. BMC Evol Biol. 2014 Mar 27;14:59. doi: 10.1186/1471-2148-14-59. BMC Evol Biol. 2014. PMID: 24669847 Free PMC article. - Environmental bacteriophages: viruses of microbes in aquatic ecosystems.
Sime-Ngando T. Sime-Ngando T. Front Microbiol. 2014 Jul 24;5:355. doi: 10.3389/fmicb.2014.00355. eCollection 2014. Front Microbiol. 2014. PMID: 25104950 Free PMC article. Review. - Diversity of Viruses Infecting the Green Microalga Ostreococcus lucimarinus.
Derelle E, Monier A, Cooke R, Worden AZ, Grimsley NH, Moreau H. Derelle E, et al. J Virol. 2015 Jun;89(11):5812-21. doi: 10.1128/JVI.00246-15. Epub 2015 Mar 18. J Virol. 2015. PMID: 25787287 Free PMC article. - Marine Prasinoviruses and Their Tiny Plankton Hosts: A Review.
Weynberg KD, Allen MJ, Wilson WH. Weynberg KD, et al. Viruses. 2017 Mar 15;9(3):43. doi: 10.3390/v9030043. Viruses. 2017. PMID: 28294997 Free PMC article. Review.
References
- Adam E, Perler FB. 2002. Development of a positive genetic selection system for inhibition of protein splicing using mycobacterial inteins in Escherichia coli DNA gyrase subunit A. J. Mol. Microbiol. Biotechnol. 4:479–488 - PubMed
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 - PubMed
- Bellec L, Grimsley N, Moreau H, Desdevises Y. 2009. Phylogenetic analysis of new prasinoviruses (Phycodnaviridae) that infect the green unicellular algae Ostreococcus, Bathycoccus and Micromonas. Environ. Microbiol. Rep. 1:114–123 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous