HBL-1 patterns synaptic remodeling in C. elegans - PubMed (original) (raw)
HBL-1 patterns synaptic remodeling in C. elegans
Katherine L Thompson-Peer et al. Neuron. 2012.
Abstract
During development, circuits are refined by the dynamic addition and removal of synapses; however, little is known about the molecular mechanisms that dictate where and when synaptic refinement occurs. Here we describe transcriptional mechanisms that pattern remodeling of C. elegans neuromuscular junctions (NMJs). The embryonic GABAergic DD motor neurons remodel their synapses, whereas the later born VD neurons do not. This specificity is mediated by differential expression of a transcription factor (HBL-1), which is expressed in DD neurons but is repressed in VDs by UNC-55/COUP-TF. DD remodeling is delayed in hbl-1 mutants whereas precocious remodeling is observed in mutants lacking the microRNA mir-84, which inhibits hbl-1 expression. Mutations increasing and decreasing circuit activity cause corresponding changes in hbl-1 expression, and corresponding shifts in the timing of DD plasticity. Thus, convergent regulation of hbl-1 expression defines a genetic mechanism that patterns activity-dependent synaptic remodeling across cell types and across developmental time.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
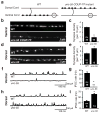
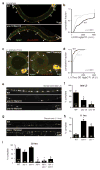
Fig. 1. Ectopic VD remodeling in unc-55 mutants
(a) Schematic illustrations of VD neuron NMJs (filled ovals) in wild type and unc-55 adults are shown. Dorsal is up and posterior is to the right in both illustrations; open circles are cell bodies. In wild type adults VD neurons retain ventral NMJs, whereas in unc-55 mutants ventral NMJs are eliminated and replaced with dorsal synapses. (b–e) Imaging of GABAergic NMJs (using the UNC-57::GFP marker). Representative images and summary data for ventral (b,c) and dorsal (d,e) GABAergic NMJs are shown for the indicated genotypes. (f–i) Representative traces and summary data for endogenous IPSCs recorded from adult ventral (f,g) and dorsal (h,i) muscles are shown for the indicated genotypes. Summary data for IPSC amplitudes are shown in Fig. S3g–h. See also figure S1.
Fig. 2. UNC-55 COUP-TF represses hbl-1 expression in VD neurons
(a–b) A representative image and summary data of hbl-1 (HgfpH, green) and GAD (red) reporter expression in wild type L3 animals. Yellow arrows indicate DD cell bodies expressing both markers, white arrows indicate DD cell bodies lacking HgfpH expression, and carrots indicate VD cell bodies. HgfpH expression was significantly lower in VD than DD neurons (p <10−5 by Kolmogorov-Smirnov test; 150 DD (black) and 260 VD (gray) cells were analyzed). (c–f) HgfpC (c,d) or HmutgfpC (e) transcriptional reporter expression (green) is compared for adjacent VD10 and DD5 neurons in wild type (c,e) and unc-55 (d) mutant animals. Gray lines connect VD10 and DD5 cells in the same animal, black lines connect median values (_p_-values by paired Student’s t test). Average log2 of the ratio of DD5 to VD10 fluorescence for HgfpC and HmutgfpC was plotted in (f), and n = number of animals analyzed (*, p<10−5 difference from WT; ns, _p_=0.2; by Student’s t test). Error bars indicate SEM. See also figure S2.
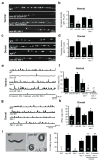
Fig. 3. Ectopic VD remodeling in unc-55 mutants requires HBL-1
(a–d) Imaging of GABAergic NMJs (using the UNC-57::GFP marker). Representative images and summary data for ventral (a,b) and dorsal (c,d) GABAergic NMJs are shown for the indicated genotypes. (e–h) Representative traces and summary data for endogenous IPSCs recorded from adult ventral (e,f) and dorsal (g,h) muscles are shown for the indicated genotypes. Summary data for IPSC amplitudes are shown in Fig. S3g–h. (i) The ventral coiling phenotype of unc-55 adults during backward locomotion is shown and quantified. The unc-55 coiling defect was partially suppressed by an hbl-1 mutation, and was restored by transgenes containing either the hbl-1 cosmid or an hbl-1 cDNA expressed by a GABA promoter (unc-25 GAD) (D neuron rescue). A single transgenic line is shown for each rescue; similar results were obtained with multiple independent transgenic lines (Fig. S3e). Error bars indicate SEM. Significant differences (p <0.01) are indicated as follows: *, significantly different from WT; #, significantly different from unc-55 single mutants; ##, significantly different from unc-55; hbl-1 double mutants. The number of animals analyzed (a–h) or replicate behavioral assays (20 animals/assay; i) is indicated for each genotype. See also figure S3.
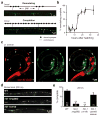
Fig. 4. DD remodeling is delayed in hbl-1 mutants
(a) DD remodeling was visualized by dorsal synapse formation with the UNC-57::GFP marker. Representative images of the dorsal cord during remodeling (above) and after completion (below) are shown. During remodeling, DD neurons form en passant synapses with the dorsal muscle. DD neuron commissures are indicated by dotted lines. (b) Summary data illustrating the time course of DD neuron remodeling is shown. 15–30 animals were analyzed for all time points except 12 hrs (where n= 160 animals). (c) The HgfpH reporter (green) is expressed in DD neurons (identified with the GAD reporter, red) during the L1 when remodeling is occurring. A representative image of a wild type L1 larva is shown. Arrows indicate the six DD cell bodies. (d and e) DD remodeling is delayed in hbl-1 mutants. Representative images of dorsal DD NMJs (d), and summary data (e) for completion of DD remodeling are shown at 23 hours post-hatching. The majority of wild-type animals have completed DD remodeling, whereas significantly fewer hbl-1 mutants have finished this process (**, p <0.0001 Chi square test). This delay was rescued by a transgene containing the hbl-1 cosmid. Error bars indicate SEM. See also figure S4.
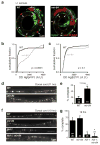
Fig. 5. The microRNA miR-84 regulates hbl-1 expression and the timing of remodeling
(a–b) Representative images (a) and summary data (b) are shown for HgfpH expression (green) in DD neurons (labeled with the GAD reporter, red, and indicated by arrows) of L1 larvae. In mir-84 mutants, HgfpH expression was significantly increased (** p <0.0001 by Kolmogorov-Smirnov test). (c) Summary data are shown comparing the fluorescent intensity of the HgfpC reporter in DD neurons of wild type and mir-84(tm1304) mutants; no significant difference was observed (p =0.1 Kolmogorov-Smirnov test). 54 wild-type and 118 mir-84 DD neurons were analyzed for median HgfpH expression (b); and 233 wild-type and 379 mir-84 DD neurons were analyzed for HgfpC expression (c). (d–e) DD remodeling occurs earlier in mir-84 mutants. Representative images (d) and summary data (e) are shown for dorsal DD NMJs at 11 hours post-hatching. Remodeling was completed significantly earlier in mir-84 mutants (*, p <0.01 Chi square test). (f–g) The impact of mir-84 on remodeling was eliminated in hbl-1; mir-84 double mutants. Representative images (f) and summary data (g) are shown for dorsal DD NMJs at 19 hours post-hatching. The extent of remodeling in hbl-1 single mutants and hbl-1; mir-84 double mutants were not significantly different. See also figure S5.
Fig. 6. Circuit activity regulates HBL-1 expression to determine the timing of DD neuron plasticity
(a–d) Representative images (a,c) and summary data (b,d) are shown for hbl-1 expression (HgfpC, green) in DD neurons (labeled with the GAD reporter, red, indicated by arrows). HgfpC expression significantly decreased in unc-13 mutants (a–b) and increased in tom-1 mutants (c–d; ** p <0.0001 Kolmogorov-Smirnov test; 72 wild type and 149 unc-13 L2 DD neurons; 64 wild type and 179 tom-1 L1 DD neurons). Expression of HgfpC in DD neurons was analyzed at different times in unc-13 and tom-1 mutants because the remodeling defects observed in these mutants occurred at different times. (e–h) DD remodeling in unc-13 and unc-18 mutants (e) or in tom-1 and slo-1 mutants (g). Representative images of dorsal GABAergic NMJs (e,g), and summary data for completion of remodeling (f,h) in late L3 animals (e,f) or at 11 hours post-hatching (g,h). (i) Summary data for completion of DD remodeling at 20 hours after hatching shows that the impact of slo-1 and tom-1 on remodeling was eliminated in double mutants with hbl-1. (* : significantly different than wild-type, p < 0.001, Chi squared test). Error bars indicate SEM, numbers indicate number of animals analyzed. See also figure S6.
Similar articles
- A transcriptional program promotes remodeling of GABAergic synapses in Caenorhabditis elegans.
Petersen SC, Watson JD, Richmond JE, Sarov M, Walthall WW, Miller DM 3rd. Petersen SC, et al. J Neurosci. 2011 Oct 26;31(43):15362-75. doi: 10.1523/JNEUROSCI.3181-11.2011. J Neurosci. 2011. PMID: 22031882 Free PMC article. - C. elegans sym-1 is a downstream target of the hunchback-like-1 developmental timing transcription factor.
Niwa R, Hada K, Moliyama K, Ohniwa RL, Tan YM, Olsson-Carter K, Chi W, Reinke V, Slack FJ. Niwa R, et al. Cell Cycle. 2009 Dec 15;8(24):4147-54. doi: 10.4161/cc.8.24.10292. Epub 2009 Dec 9. Cell Cycle. 2009. PMID: 19923914 Free PMC article. - Synaptic remodeling, lessons from C. elegans.
Cuentas-Condori A, Miller Rd DM. Cuentas-Condori A, et al. J Neurogenet. 2020 Sep-Dec;34(3-4):307-322. doi: 10.1080/01677063.2020.1802725. Epub 2020 Aug 18. J Neurogenet. 2020. PMID: 32808848 Free PMC article. - Transcriptional Control of Synaptic Remodeling through Regulated Expression of an Immunoglobulin Superfamily Protein.
He S, Philbrook A, McWhirter R, Gabel CV, Taub DG, Carter MH, Hanna IM, Francis MM, Miller DM 3rd. He S, et al. Curr Biol. 2015 Oct 5;25(19):2541-8. doi: 10.1016/j.cub.2015.08.022. Epub 2015 Sep 17. Curr Biol. 2015. PMID: 26387713 Free PMC article. - Meis/UNC-62 isoform dependent regulation of CoupTF-II/UNC-55 and GABAergic motor neuron subtype differentiation.
Campbell RF, Walthall WW. Campbell RF, et al. Dev Biol. 2016 Nov 15;419(2):250-261. doi: 10.1016/j.ydbio.2016.09.009. Epub 2016 Sep 12. Dev Biol. 2016. PMID: 27634571
Cited by
- Neural Circuit Remodeling: Mechanistic Insights from Invertebrates.
Liu S, Alexander KD, Francis MM. Liu S, et al. J Dev Biol. 2024 Oct 11;12(4):27. doi: 10.3390/jdb12040027. J Dev Biol. 2024. PMID: 39449319 Free PMC article. Review. - The DEG/ENaC cation channel protein UNC-8 drives activity-dependent synapse removal in remodeling GABAergic neurons.
Miller-Fleming TW, Petersen SC, Manning L, Matthewman C, Gornet M, Beers A, Hori S, Mitani S, Bianchi L, Richmond J, Miller DM. Miller-Fleming TW, et al. Elife. 2016 Jul 12;5:e14599. doi: 10.7554/eLife.14599. Elife. 2016. PMID: 27403890 Free PMC article. - Synaptogenesis: unmasking molecular mechanisms using Caenorhabditis elegans.
Mizumoto K, Jin Y, Bessereau JL. Mizumoto K, et al. Genetics. 2023 Feb 9;223(2):iyac176. doi: 10.1093/genetics/iyac176. Genetics. 2023. PMID: 36630525 Free PMC article. - The homeodomain transcriptional regulator DVE-1 directs a program for synapse elimination during circuit remodeling.
Alexander KD, Ramachandran S, Biswas K, Lambert CM, Russell J, Oliver DB, Armstrong W, Rettler M, Liu S, Doitsidou M, Bénard C, Walker AK, Francis MM. Alexander KD, et al. Nat Commun. 2023 Nov 18;14(1):7520. doi: 10.1038/s41467-023-43281-4. Nat Commun. 2023. PMID: 37980357 Free PMC article. - Gut neuroendocrine signaling regulates synaptic assembly in C. elegans.
Shi Y, Qin L, Wu M, Zheng J, Xie T, Shao Z. Shi Y, et al. EMBO Rep. 2022 Aug 3;23(8):e53267. doi: 10.15252/embr.202153267. Epub 2022 Jun 24. EMBO Rep. 2022. PMID: 35748387 Free PMC article.
References
- Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Developmental Cell. 2003;4:625–637. - PubMed
- Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. - PubMed
- Ambros V, Horvitz HR. The lin-14 locus of Caenorhabditis elegans controls the time of expression of specific postembryonic developmental events. Genes & Development. 1987;1:398–414. - PubMed
- Armentano M, Chou SJ, (null) (null), O’Leary DDM, Studer M. COUP-TFI regulates the balance of cortical patterning between frontal/motor and sensory areas. Nat Neurosci. 2007;10:1277–1286. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 NS032196-17S1/NS/NINDS NIH HHS/United States
- R37 NS032196-19/NS/NINDS NIH HHS/United States
- R01 NS032196/NS/NINDS NIH HHS/United States
- K99MH085039/MH/NIMH NIH HHS/United States
- R00 MH085039/MH/NIMH NIH HHS/United States
- R37 NS032196-16/NS/NINDS NIH HHS/United States
- R37 NS032196-17/NS/NINDS NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- NS32196/NS/NINDS NIH HHS/United States
- R37 NS032196/NS/NINDS NIH HHS/United States
- R37 NS032196-18/NS/NINDS NIH HHS/United States
- K99 MH085039/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous