Paternal age effect mutations and selfish spermatogonial selection: causes and consequences for human disease - PubMed (original) (raw)
Review
Paternal age effect mutations and selfish spermatogonial selection: causes and consequences for human disease
Anne Goriely et al. Am J Hum Genet. 2012.
Abstract
Advanced paternal age has been associated with an increased risk for spontaneous congenital disorders and common complex diseases (such as some cancers, schizophrenia, and autism), but the mechanisms that mediate this effect have been poorly understood. A small group of disorders, including Apert syndrome (caused by FGFR2 mutations), achondroplasia, and thanatophoric dysplasia (FGFR3), and Costello syndrome (HRAS), which we collectively term "paternal age effect" (PAE) disorders, provides a good model to study the biological and molecular basis of this phenomenon. Recent evidence from direct quantification of PAE mutations in sperm and testes suggests that the common factor in the paternal age effect lies in the dysregulation of spermatogonial cell behavior, an effect mediated molecularly through the growth factor receptor-RAS signal transduction pathway. The data show that PAE mutations, although arising rarely, are positively selected and expand clonally in normal testes through a process akin to oncogenesis. This clonal expansion, which is likely to take place in the testes of all men, leads to the relative enrichment of mutant sperm over time-explaining the observed paternal age effect associated with these disorders-and in rare cases to the formation of testicular tumors. As regulation of RAS and other mediators of cellular proliferation and survival is important in many different biological contexts, for example during tumorigenesis, organ homeostasis and neurogenesis, the consequences of selfish mutations that hijack this process within the testis are likely to extend far beyond congenital skeletal disorders to include complex diseases, such as neurocognitive disorders and cancer predisposition.
Copyright © 2012 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures
Figure 1
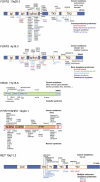
Rate of Germline Mutations in the Human Genome on a Logarithmic Scale (A) Background rate of nucleotide substitution quantified by direct measurements of transgenerational mutational load (unfilled inverted triangles in dark gray box) and indirect estimates (lighter gray box) for all substitutions (inverted triangles), transitions at CpG dinucleotides (squares), transversions at CpG dinucleotides (circles), transitions at non-CpG dinucleotides (diamonds), and transversions at non-CpG dinucleotides (triangles). Data source indicated by symbol shading as follows: black, gray, and white. (B) Estimated birth prevalence for de novo mutations in two PAE syndromes (green box). Apert syndrome FGFR2 mutations caused by c.755C>G transversion at a CpG dinucleotide account for 66% of cases (red circle) and mutations caused by c.758C>G transversion at a non-CpG dinucleotide for 33% of cases (red triangle). Achondroplasia FGFR3 mutations caused by c.1138G>A transition or c.1138G>C transversion at a CpG dinucleotide are marked by the green square and circle, respectively. Note that these figures assume that all mutations are paternal in origin. (C) Direct measurements of mutation levels in human sperm at specific locations in the genome. Symbols for each mutation (square, circle, diamond, and triangle) refer to mutation types as in (A). Top blue boxes show average levels for the FGFR2 c.752–755 positions (encompassing codons p.Arg251 and p.Ser252) measured by RED/PCR/pyrosequencing in the sperm of 99 healthy men. The strongly activating c.755C>G (p.Ser252Trp) Apert transversion (red circle in dark blue box) and the c.755C>T (p.Ser252Leu) transition (yellow square in dark blue box) are associated with gain-of-function (GOF) properties; these two substitutions are found at significantly higher levels in sperm than any of the ten other mutations encoding silent or loss-of-function (LOF) mutations (light blue box). The average age of the sperm donors was 37.4 years. Middle blue box shows average levels of the two Apert mutations measured in the sperm of 323 healthy men by Bi-PAP-A. The average age of the sperm donors was 38.7 years. Note that the very similar estimate for the FGFR2 mutation c.755C>G (red circles) was obtained by different methods. Prevalence in sperm exceeds birth prevalence (B) because sperm donors were older on average than the population of fathers. Bottom blue boxes show average levels at the FGFR3 codon p.Lys650 in the sperm of 78 men via RED/PCR/massively parallel sequencing. The substitutions encoding proteins with gain-of-function properties (dark blue box), and in particular the c.1948A>G transition (p.Lys650Glu, causing TDII, green diamond), are found at higher levels in sperm than the changes associated with silent/loss-of-function substitutions (light blue box). The average age of the sperm donors was 40.2 years.
Figure 2
Relative Prevalence of FGFR2 and FGFR3 Mutations Explained by Combined Effects of Copy Error, at the DNA level, and Selfish Selection, at the Protein Level, in SSC (A) Mutational events taking place at FGFR2 position c.755C. Each pair of lines with gray boxes denotes the two FGFR alleles (box represents the exon and lines represent the intron. Asterisks in (A) represent a SNP located in the intron upstream of FGFR2 position c.755; blue and purple denote the two different alleles of the SNP. Circles and squares represent single events of transversion and transition at CpG dinucleotide sites, respectively. Triangular sectors indicate expansion over time of clones carrying the resulting mutant protein (selection). wt is used as an abbreviation for wild-type. The c.755C>G transversion (red circle) occurs rarely, but the resulting p.Ser252Trp substitution (causing Apert syndrome in the germline) confers a strong selective advantage to the mutant SSC (red triangular sector), leading to clonal expansion over time. The background mutation rate for the c.755C>T transition at this CpG dinucleotide (yellow square) is higher (∼3.3- to 10-fold), but the resulting mutant protein (p.Ser252Leu associated with Crouzon syndrome) confers a weaker selective advantage compared to p.Ser252Trp, leading to slower clonal expansion (yellow triangular sectors). The original mutation events occur randomly on either FGFR2 allele (blue or purple asterisk in men heterozygous for the SNP) at different times within the testis of an aging man. (B) Accumulation of c.755C mutations in FGFR2 in sperm over time. Because of the rarity of the mutational events, the distribution of the linked SNP (graphs, blue and purple columns on the right) is more skewed for the c.755C>G (above) than the c.755C>T mutation (below); however, the prevalence of c.755C>G is greater than c.755C>T because of the greater selective advantage conferred by the p.Ser252Trp compared to the p.Ser252Leu mutant protein. (C) Mutational events taking place at FGFR3 position c.1138G cause achondroplasia. The c.1138G>A transition (green square) is more frequent than the c.1138G>C transversion (light green circle) at this CpG dinucleotide site. Both resulting proteins encode p.Gly380Arg that confers the same selective advantage to the mutant SSC (green triangular sectors). The difference in birth prevalence of achondroplasia caused by the two mutations is likely to be explained by the relative frequency of the original mutational events at FGFR3 position c.1138. Although selection for the FGFR3 achondroplasia mutation is probably weaker than for the FGFR2 Apert mutation, nevertheless achondroplasia mutations occur more frequently because of the high intrinsic rate of the c.1138G>A transition.
Figure 3
Somatic and Germline Gain-Of-Function Mutations Associated with the Five PAE Genes In each case, the name of the gene or protein, its genomic location, and the functional protein domains (see abbreviations below) are indicated; the most common mutations (single-letter amino acid codes) associated with cancer (COSMIC database) are indicated above the schematic of the protein along with some of the cancers for which the mutations have been described. The germline mutations associated with congenital syndromes (Human Gene Mutation Database [HGMD] database) are indicated below the protein and are color coded according to the associated disorders (key on the figure). A few complex rearrangements and splice-site mutations are omitted for clarity. Proteins are numbered and drawn to scale (except for HRAS, for which the scale is 3:1, and RET, for which the scale is 0.9:1) according to the reference sequences (for RefSeq, see footnote b in Table 1); the length in amino acids (AA) is indicated in bold. The following abbreviations are used: Ig, immunoglobulin-like domain; TM, transmembrane domain; TK, tyrosine kinase domain; P loop, phosphate-binding loop; Switch, switch domain; CVLS, prenylation signal sequence; SH2, Src Homology 2 domain; PTP, phosphotyrosine domain; CAD, cadherin domain; CRD, cysteine-rich domain; SADDAN, severe achondroplasia with developmental delay and acanthosis nigricans; AN, acanthosis nigricans; and JMML, juvenile myelomonocytic leukemia.
Figure 4
PAE Disorders Cluster within the Receptor Tyrosine Kinase (RTK)-RAS Signaling Pathway The known PAE disorders (boxes) and the five PAE proteins that fulfill all three criteria to belong to the PAE class (as defined in the text) are indicated in blue. Other candidate PAE disorders (Table 2) are in black within the boxes. Signaling downstream of RAS involves the RAF/MEK/ERK (MAPK-ERK branch) (in red) and the PI3K-AKT-mTOR (in green) pathways. The MEK1/2 inhibitor (in red box) specifically blocks the phosphorylation of ERK1/2. For details and abbreviations see the main text.
Figure 5
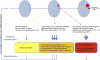
Long-Term Consequences of Selfish Selection in the Testis Blue ovals represent the testis at three ages from puberty (left) to senescence (right). During the recurrent rounds of replication required for spermatogenesis and SSC self-renewal, stochastic mutations (represented by X) occur randomly in the testis. Depending on the functional consequence of the resulting mutation for the SSC, three scenarios are illustrated. Functionally neutral mutations (in yellow; left part of each testis) do not accumulate and are associated with a very low risk of individual transmission (∼10−8, the background rate of nucleotide substitution in the genome). In red (right part of the testis) are typical PAE mutations that confer a strong selective advantage to the mutant SSC, leading over time to the formation of large “mole-like” clones (ovals) and an increased risk of transmission in older men (up to 1,000-fold higher than the background mutation rate). In rare cases, these mutations are associated with spermatocytic seminoma (SPS). In orange (bottom part of the testis) is depicted an intermediate scenario for mutations with milder selective advantage (i.e., weak gain-of-function, change in copy number or regulation of expression) that are enriched over time in SSC to a lesser extent (>1- to 100-fold). Many such mutational targets could potentially exist. Although strongly activating mutations associated with classical PAE disorders are deleterious and will be rapidly eliminated because of low reproductive fitness, neutral and mildly pathogenic mutations are potentially transmissible over many generations, contributing to genetic heterogeneity. Some mildly pathogenic mutations, although associated with deleterious phenotypes, might be maintained in the population either by recurrent mutation or because they provide a beneficial fitness trait during spermatogenesis.
Similar articles
- Selfish spermatogonial selection: evidence from an immunohistochemical screen in testes of elderly men.
Lim J, Maher GJ, Turner GD, Dudka-Ruszkowska W, Taylor S, Rajpert-De Meyts E, Goriely A, Wilkie AO. Lim J, et al. PLoS One. 2012;7(8):e42382. doi: 10.1371/journal.pone.0042382. Epub 2012 Aug 6. PLoS One. 2012. PMID: 22879958 Free PMC article. - Cellular evidence for selfish spermatogonial selection in aged human testes.
Maher GJ, Goriely A, Wilkie AO. Maher GJ, et al. Andrology. 2014 May;2(3):304-14. doi: 10.1111/j.2047-2927.2013.00175.x. Epub 2013 Dec 19. Andrology. 2014. PMID: 24357637 Review. - Selfish mutations dysregulating RAS-MAPK signaling are pervasive in aged human testes.
Maher GJ, Ralph HK, Ding Z, Koelling N, Mlcochova H, Giannoulatou E, Dhami P, Paul DS, Stricker SH, Beck S, McVean G, Wilkie AOM, Goriely A. Maher GJ, et al. Genome Res. 2018 Dec;28(12):1779-1790. doi: 10.1101/gr.239186.118. Epub 2018 Oct 24. Genome Res. 2018. PMID: 30355600 Free PMC article. - Functional robustness of adult spermatogonial stem cells after induction of hyperactive Hras.
Yamada M, Cai W, Martin LA, N'Tumba-Byn T, Seandel M. Yamada M, et al. PLoS Genet. 2019 May 3;15(5):e1008139. doi: 10.1371/journal.pgen.1008139. eCollection 2019 May. PLoS Genet. 2019. PMID: 31050682 Free PMC article. - "Selfish spermatogonial selection": a novel mechanism for the association between advanced paternal age and neurodevelopmental disorders.
Goriely A, McGrath JJ, Hultman CM, Wilkie AO, Malaspina D. Goriely A, et al. Am J Psychiatry. 2013 Jun;170(6):599-608. doi: 10.1176/appi.ajp.2013.12101352. Am J Psychiatry. 2013. PMID: 23639989 Free PMC article. Review.
Cited by
- Advances in Skeletal Dysplasia Genetics.
Geister KA, Camper SA. Geister KA, et al. Annu Rev Genomics Hum Genet. 2015;16:199-227. doi: 10.1146/annurev-genom-090314-045904. Epub 2015 Apr 22. Annu Rev Genomics Hum Genet. 2015. PMID: 25939055 Free PMC article. Review. - Selfish spermatogonial selection: evidence from an immunohistochemical screen in testes of elderly men.
Lim J, Maher GJ, Turner GD, Dudka-Ruszkowska W, Taylor S, Rajpert-De Meyts E, Goriely A, Wilkie AO. Lim J, et al. PLoS One. 2012;7(8):e42382. doi: 10.1371/journal.pone.0042382. Epub 2012 Aug 6. PLoS One. 2012. PMID: 22879958 Free PMC article. - Measurement of FGFR3 signaling at the cell membrane via total internal reflection fluorescence microscopy to compare the activation of FGFR3 mutants.
Hartl I, Brumovska V, Striedner Y, Yasari A, Schütz GJ, Sevcsik E, Tiemann-Boege I. Hartl I, et al. J Biol Chem. 2023 Feb;299(2):102832. doi: 10.1016/j.jbc.2022.102832. Epub 2022 Dec 27. J Biol Chem. 2023. PMID: 36581204 Free PMC article. - RASopathies: From germline mutations to somatic and multigenic diseases.
Riller Q, Rieux-Laucat F. Riller Q, et al. Biomed J. 2021 Aug;44(4):422-432. doi: 10.1016/j.bj.2021.06.004. Epub 2021 Jun 24. Biomed J. 2021. PMID: 34175492 Free PMC article. Review. - The Age of the Father.
Poot M. Poot M. Mol Syndromol. 2017 Jun;8(4):169-171. doi: 10.1159/000471776. Epub 2017 Apr 20. Mol Syndromol. 2017. PMID: 28690481 Free PMC article. No abstract available.
References
- Green R.F., Devine O., Crider K.S., Olney R.S., Archer N., Olshan A.F., Shapira S.K., National Birth Defects Prevention Study Association of paternal age and risk for major congenital anomalies from the National Birth Defects Prevention Study, 1997 to 2004. Ann. Epidemiol. 2010;20:241–249. - PMC - PubMed
- Yip B.H., Pawitan Y., Czene K. Parental age and risk of childhood cancers: A population-based cohort study from Sweden. Int. J. Epidemiol. 2006;35:1495–1503. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous