Outcome of the first electron microscopy validation task force meeting - PubMed (original) (raw)
. 2012 Feb 8;20(2):205-14.
doi: 10.1016/j.str.2011.12.014.
Andrej Sali, Matthew L Baker, Bridget Carragher, Batsal Devkota, Kenneth H Downing, Edward H Egelman, Zukang Feng, Joachim Frank, Nikolaus Grigorieff, Wen Jiang, Steven J Ludtke, Ohad Medalia, Pawel A Penczek, Peter B Rosenthal, Michael G Rossmann, Michael F Schmid, Gunnar F Schröder, Alasdair C Steven, David L Stokes, John D Westbrook, Willy Wriggers, Huanwang Yang, Jasmine Young, Helen M Berman, Wah Chiu, Gerard J Kleywegt, Catherine L Lawson
Affiliations
- PMID: 22325770
- PMCID: PMC3328769
- DOI: 10.1016/j.str.2011.12.014
Outcome of the first electron microscopy validation task force meeting
Richard Henderson et al. Structure. 2012.
Abstract
This Meeting Review describes the proceedings and conclusions from the inaugural meeting of the Electron Microscopy Validation Task Force organized by the Unified Data Resource for 3DEM (http://www.emdatabank.org) and held at Rutgers University in New Brunswick, NJ on September 28 and 29, 2010. At the workshop, a group of scientists involved in collecting electron microscopy data, using the data to determine three-dimensional electron microscopy (3DEM) density maps, and building molecular models into the maps explored how to assess maps, models, and other data that are deposited into the Electron Microscopy Data Bank and Protein Data Bank public data archives. The specific recommendations resulting from the workshop aim to increase the impact of 3DEM in biology and medicine.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures
Figure 1
EMDataBank, Unified Data Resource for 3DEM Home Page Available at
.
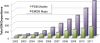
Figure 2
3DEM Entries in EMDB and PDB, Cumulative by Year Statistics for December 31, 2011: 1322 map entries, 427 model entries.
Similar articles
- EMDataBank unified data resource for 3DEM.
Lawson CL, Patwardhan A, Baker ML, Hryc C, Garcia ES, Hudson BP, Lagerstedt I, Ludtke SJ, Pintilie G, Sala R, Westbrook JD, Berman HM, Kleywegt GJ, Chiu W. Lawson CL, et al. Nucleic Acids Res. 2016 Jan 4;44(D1):D396-403. doi: 10.1093/nar/gkv1126. Epub 2015 Nov 17. Nucleic Acids Res. 2016. PMID: 26578576 Free PMC article. - EMDataBank.org: unified data resource for CryoEM.
Lawson CL, Baker ML, Best C, Bi C, Dougherty M, Feng P, van Ginkel G, Devkota B, Lagerstedt I, Ludtke SJ, Newman RH, Oldfield TJ, Rees I, Sahni G, Sala R, Velankar S, Warren J, Westbrook JD, Henrick K, Kleywegt GJ, Berman HM, Chiu W. Lawson CL, et al. Nucleic Acids Res. 2011 Jan;39(Database issue):D456-64. doi: 10.1093/nar/gkq880. Epub 2010 Oct 8. Nucleic Acids Res. 2011. PMID: 20935055 Free PMC article. - EMDB-the Electron Microscopy Data Bank.
wwPDB Consortium. wwPDB Consortium. Nucleic Acids Res. 2024 Jan 5;52(D1):D456-D465. doi: 10.1093/nar/gkad1019. Nucleic Acids Res. 2024. PMID: 37994703 Free PMC article. Review. - Web-based visualisation and analysis of 3D electron-microscopy data from EMDB and PDB.
Lagerstedt I, Moore WJ, Patwardhan A, Sanz-García E, Best C, Swedlow JR, Kleywegt GJ. Lagerstedt I, et al. J Struct Biol. 2013 Nov;184(2):173-81. doi: 10.1016/j.jsb.2013.09.021. Epub 2013 Oct 7. J Struct Biol. 2013. PMID: 24113529 Free PMC article. - The Protein Data Bank Archive.
Velankar S, Burley SK, Kurisu G, Hoch JC, Markley JL. Velankar S, et al. Methods Mol Biol. 2021;2305:3-21. doi: 10.1007/978-1-0716-1406-8_1. Methods Mol Biol. 2021. PMID: 33950382 Review.
Cited by
- Validated near-atomic resolution structure of bacteriophage epsilon15 derived from cryo-EM and modeling.
Baker ML, Hryc CF, Zhang Q, Wu W, Jakana J, Haase-Pettingell C, Afonine PV, Adams PD, King JA, Jiang W, Chiu W. Baker ML, et al. Proc Natl Acad Sci U S A. 2013 Jul 23;110(30):12301-6. doi: 10.1073/pnas.1309947110. Epub 2013 Jul 9. Proc Natl Acad Sci U S A. 2013. PMID: 23840063 Free PMC article. - Cryo-EM structure of a novel calicivirus, Tulane virus.
Yu G, Zhang D, Guo F, Tan M, Jiang X, Jiang W. Yu G, et al. PLoS One. 2013;8(3):e59817. doi: 10.1371/journal.pone.0059817. Epub 2013 Mar 22. PLoS One. 2013. PMID: 23533651 Free PMC article. - Native structure of a retroviral envelope protein and its conformational change upon interaction with the target cell.
Riedel C, Vasishtan D, Siebert CA, Whittle C, Lehmann MJ, Mothes W, Grünewald K. Riedel C, et al. J Struct Biol. 2017 Feb;197(2):172-180. doi: 10.1016/j.jsb.2016.06.017. Epub 2016 Jun 23. J Struct Biol. 2017. PMID: 27345930 Free PMC article. - CueR activates transcription through a DNA distortion mechanism.
Fang C, Philips SJ, Wu X, Chen K, Shi J, Shen L, Xu J, Feng Y, O'Halloran TV, Zhang Y. Fang C, et al. Nat Chem Biol. 2021 Jan;17(1):57-64. doi: 10.1038/s41589-020-00653-x. Epub 2020 Sep 28. Nat Chem Biol. 2021. PMID: 32989300 Free PMC article. - Structure of the bacterial ribosome at 2 Å resolution.
Watson ZL, Ward FR, Méheust R, Ad O, Schepartz A, Banfield JF, Cate JH. Watson ZL, et al. Elife. 2020 Sep 14;9:e60482. doi: 10.7554/eLife.60482. Elife. 2020. PMID: 32924932 Free PMC article.
References
- Agard D.A. J. Mol. Biol. 1983;167:849–852. - PubMed
- Al-Amoudi A., Norlen L.P., Dubochet J. J. Struct. Biol. 2004;148:131–135. - PubMed
- Al-Amoudi A., Díez D.C., Betts M.J., Frangakis A.S. Nature. 2007;450:832–837. - PubMed
- Alber F., Förster F., Korkin D., Topf M., Sali A. Annu. Rev. Biochem. 2008;77:443–477. - PubMed
- Baker M.L., Jiang W., Bowman B.R., Zhou Z.H., Quiocho F.A., Rixon F.J., Chiu W. J. Mol. Biol. 2003;331:447–456. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P41RR002250/RR/NCRR NIH HHS/United States
- MC_U105184322/MRC_/Medical Research Council/United Kingdom
- R01 GM079429/GM/NIGMS NIH HHS/United States
- MC_U117581334/MRC_/Medical Research Council/United Kingdom
- R01 GM060635/GM/NIGMS NIH HHS/United States
- P41 RR002250/RR/NCRR NIH HHS/United States
- R01GM079429/GM/NIGMS NIH HHS/United States
- BB/G022577/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 088944/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources