Iterative stable alignment and clustering of 2D transmission electron microscope images - PubMed (original) (raw)
Iterative stable alignment and clustering of 2D transmission electron microscope images
Zhengfan Yang et al. Structure. 2012.
Abstract
Identification of homogeneous subsets of images in a macromolecular electron microscopy (EM) image data set is a critical step in single-particle analysis. The task is handled by iterative algorithms, whose performance is compromised by the compounded limitations of image alignment and K-means clustering. Here we describe an approach, iterative stable alignment and clustering (ISAC) that, relying on a new clustering method and on the concepts of stability and reproducibility, can extract validated, homogeneous subsets of images. ISAC requires only a small number of simple parameters and, with minimal human intervention, can eliminate bias from two-dimensional image clustering and maximize the quality of group averages that can be used for ab initio three-dimensional structural determination and analysis of macromolecular conformational variability. Repeated testing of the stability and reproducibility of a solution within ISAC eliminates heterogeneous or incorrect classes and introduces critical validation to the process of EM image clustering.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures
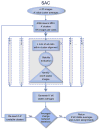
Figure 1. Flowchart of Stable Alignment and Clustering
Only clusters comprising images with alignment parameters that are stable (at a given pixel error threshold) across several independent rounds of within-cluster alignment are retained. Images in unstable clusters are sent back to the unassigned image pool for reclustering. See also Figure S5.
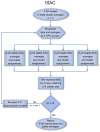
Figure 2. Flowchart of Iterative Stable Alignment and Clustering
The membership of clusters generated by four semi-independent SAC runs is compared, and only clusters with reproducible membership are retained. Images in clusters with low reproducibility are sent back to the unassigned image pool for reclustering.
Figure 3. ISAC Results Obtained for the Data Set of EF-Tu Ribosomal Complex
(A) Raw EM images. (B) Selection of ISAC cluster averages matched to projections of the X-ray structure. (C) Common lines volume compared with a map derived from the X-ray model. Scale bar corresponds to 10 nm. See also Figures S1–S3 and S6–S11.
Figure 4. ISAC Analysis of hRNAPII EM Images
(A) Raw EM images of hRNAPII. (B) A selection of hRNAPII ISAC cluster averages matched to projections of the X-ray structure of yeast RNAPII (pdb 1WCM). (C) A 3D map of hRNAPII derived by applying common lines to the ISAC averages shown in (B) (left), compared to a map of the homologous yeast RNAPII (right) derived from its X-ray structure. Scale bars correspond to 10 nm in (A and B) and 5 nm in (C). See also Figure S4.
Figure 5. Conformational Variability of hRNAPII Revealed by ISAC Cluster Averages
(A) Selected hRNAPII ISAC averages showing changes in the position/ appearance of the clamp domain (marked by yellow arrowheads). (B) hRNAPII volumes obtained after CD-PCA analysis of resampled ISAC averages show variability in clamp structure. (C) Two 3D maps obtained by competitive refinement of hRNAPII images using the volumes in (B) as initial references show alternative conformations of the hRNAPII clamp domain. Scale bars correspond to 10 nm in (A) and 5 nm in (B and C).
Similar articles
- A Stochastic Hill Climbing Approach for Simultaneous 2D Alignment and Clustering of Cryogenic Electron Microscopy Images.
Reboul CF, Bonnet F, Elmlund D, Elmlund H. Reboul CF, et al. Structure. 2016 Jun 7;24(6):988-96. doi: 10.1016/j.str.2016.04.006. Epub 2016 May 12. Structure. 2016. PMID: 27184214 - Ab initio structure determination from electron microscopic images of single molecules coexisting in different functional states.
Elmlund D, Davis R, Elmlund H. Elmlund D, et al. Structure. 2010 Jul 14;18(7):777-86. doi: 10.1016/j.str.2010.06.001. Structure. 2010. PMID: 20637414 - A Fast Image Alignment Approach for 2D Classification of Cryo-EM Images Using Spectral Clustering.
Wang X, Lu Y, Liu J. Wang X, et al. Curr Issues Mol Biol. 2021 Oct 18;43(3):1652-1668. doi: 10.3390/cimb43030117. Curr Issues Mol Biol. 2021. PMID: 34698131 Free PMC article. - Advances in image processing for single-particle analysis by electron cryomicroscopy and challenges ahead.
Vilas JL, Tabassum N, Mota J, Maluenda D, Jiménez-Moreno A, Majtner T, Carazo JM, Acton ST, Sorzano COS. Vilas JL, et al. Curr Opin Struct Biol. 2018 Oct;52:127-145. doi: 10.1016/j.sbi.2018.11.004. Epub 2018 Nov 30. Curr Opin Struct Biol. 2018. PMID: 30509756 Review. - [A review of automatic particle recognition in Cryo-EM images].
Wu X, Wu X. Wu X, et al. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2010 Oct;27(5):1178-82. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2010. PMID: 21089695 Review. Chinese.
Cited by
- Comparative Structural and Computational Analysis Supports Eighteen Cellulose Synthases in the Plant Cellulose Synthesis Complex.
Nixon BT, Mansouri K, Singh A, Du J, Davis JK, Lee JG, Slabaugh E, Vandavasi VG, O'Neill H, Roberts EM, Roberts AW, Yingling YG, Haigler CH. Nixon BT, et al. Sci Rep. 2016 Jun 27;6:28696. doi: 10.1038/srep28696. Sci Rep. 2016. PMID: 27345599 Free PMC article. - Pre-pro is a fast pre-processor for single-particle cryo-EM by enhancing 2D classification.
Chung SC, Lin HH, Niu PY, Huang SH, Tu IP, Chang WH. Chung SC, et al. Commun Biol. 2020 Sep 11;3(1):508. doi: 10.1038/s42003-020-01229-0. Commun Biol. 2020. PMID: 32917929 Free PMC article. - Membrane Protein Complex ExbB4-ExbD1-TonB1 from Escherichia coli Demonstrates Conformational Plasticity.
Sverzhinsky A, Chung JW, Deme JC, Fabre L, Levey KT, Plesa M, Carter DM, Lypaczewski P, Coulton JW. Sverzhinsky A, et al. J Bacteriol. 2015 Jun;197(11):1873-85. doi: 10.1128/JB.00069-15. Epub 2015 Mar 23. J Bacteriol. 2015. PMID: 25802296 Free PMC article. - Glycan-dependent cell adhesion mechanism of Tc toxins.
Roderer D, Bröcker F, Sitsel O, Kaplonek P, Leidreiter F, Seeberger PH, Raunser S. Roderer D, et al. Nat Commun. 2020 Jun 1;11(1):2694. doi: 10.1038/s41467-020-16536-7. Nat Commun. 2020. PMID: 32483155 Free PMC article. - Structure, mechanism, and regulation of the chloroplast ATP synthase.
Hahn A, Vonck J, Mills DJ, Meier T, Kühlbrandt W. Hahn A, et al. Science. 2018 May 11;360(6389):eaat4318. doi: 10.1126/science.aat4318. Science. 2018. PMID: 29748256 Free PMC article.
References
- Armache KJ, Mitterweger S, Meinhart A, Cramer P. Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J Biol Chem. 2005;280:7131–7134. - PubMed
- Baldwin PR, Penczek PA. The transform class in SPARX and EMAN2. J Struct Biol. 2007;157:250–261. - PubMed
- Burkard RE, Mauro D, Martello S. Assignment Problems. Philadelphia: Society for Industrial and Applied Mathematics; 2009.
- Duda RO, Hart PE, Stork DG. Pattern Classification. New York: Wiley; 2001.
- Grundel DA, Pardalos PM. Test problem generator for the multidimensional assignment problem. Comput Optim Appl. 2005;30:133–146.
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM067167/GM/NIGMS NIH HHS/United States
- R01 GM60635/GM/NIGMS NIH HHS/United States
- R01 GM060635/GM/NIGMS NIH HHS/United States
- R01 GM060635-14/GM/NIGMS NIH HHS/United States
- R01 GM067167-08/GM/NIGMS NIH HHS/United States
- R01 GM67167/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources