Frequency of epitope-specific naive CD4(+) T cells correlates with immunodominance in the human memory repertoire - PubMed (original) (raw)
Frequency of epitope-specific naive CD4(+) T cells correlates with immunodominance in the human memory repertoire
William W Kwok et al. J Immunol. 2012.
Abstract
The frequency of epitope-specific naive CD4(+) T cells in humans has not been extensively examined. In this study, a systematic approach was used to examine the frequency of CD4(+) T cells that recognize the protective Ag of Bacillus anthracis in both anthrax vaccine-adsorbed vaccinees and nonvaccinees with HLA-DRB1*01:01 haplotypes. Three epitopes were identified that had distinct degrees of immunodominance in subjects that had received the vaccine. Average naive precursor frequencies of T cells specific for these different epitopes in the human repertoire ranged from 0.2 to 10 per million naive CD4(+) T cells, which is comparable to precursor frequencies observed in the murine repertoire. Frequencies of protective Ag-specific T cells were two orders of magnitude higher in immunized subjects than in nonvaccinees. The frequencies of epitope-specific memory CD4(+) T cells in vaccinees were directly correlated with the frequencies of precursors in the naive repertoire. At the level of TCR usage, at least one preferred Vβ in the naive repertoire was present in the memory repertoire. These findings implicate naive frequencies as a crucial factor in shaping the epitope specificity of memory CD4(+) T cell responses.
Figures
Figure 1
Frequencies and phenotypes of DR0101 restricted PA reactive T cells in DR0101 AVA vaccinees. A. PBMC from a DR0101 subject were stained with DR0101/PA401-420, DR0101/PA505-524 and DR0101/PA713-732 PE-tetramers and anti-CD45RA antibody, and with gating antibodies ex vivo. The cells were subsequently incubated with anti-PE magnetic beads and enriched with a magnetic column. Frequencies of PA specific T cells were determined by dividing the number of tetramer positive cells in the enriched fraction by the total number of CD4+ T cells in the sample before fractionation, determined to be 63, 22 and 1,000 per million total CD4+ T cells for PA401-420, PA505-524 and PA713-732 epitopes, respectively. B. Frequencies of PA401-420, PA505-524 and PA713-732 T cells in four different DR0101 AVA vaccinees. Each symbol represents the epitope specific T cell frequency for a single subject. The p values were determined by using two-tailed unpaired t tests. C. CD27 and CCR4 expression of PA713-732 reactive CD4+ T cells. For each panel (A-C), we gated on CD4+, CD14-, CD19- and Via-Probe negative cells. The percentages of tetramer positive cells that expressed the surface marker are as indicated. Data as shown are representative results from two or more experiments in each of the 4 subjects.
Figure 2
Frequencies and phenotypes of PA713-732 reactive T cells in non-AVA vaccines. A. PBMC from a DR0101 subject were stained with DR0101/PA713-732 PE-tetramers, CD4 antibody, and gating antibodies (anti-CD3, anti-CD14 and anti-CD19) ex vivo. B. PBMC were stained with DR0101/PA713-732 PE-tetramers, CD8 antibody, and gating antibodies ex vivo. Samples from A and B were obtained from the same subject and the frequency was determined to be 6 per million total CD4+ T cells. C. PBMC from a second subject were co-stained with DR0101/PA713-732 tetramers labeled with either PE or allophycocyanin and stained with gating antibodies (anti-CD4, anti-CD14 and anti-CD19) ex vivo. The frequency was determined to be 3 per million total CD4+ T cells. D. E and F. PBMC from DR0101 subjects were stained with DR0101/PA713-732 PE-tetramers, gating antibodies and anti-CD45RA, anti-CCR4 and anti-CD27 antibodies ex vivo. For each panel (A-F), we enriched using anti-PE magnetic beads and examined tetramer positive cells ex vivo, gating on forward and side scattering, and then on cells that were Via-Probe-, CD3+ or CD4+ , CD14- and CD19-. The percentages of tetramer positive cells that expressed the surface marker are as indicated. Data shown for each panel are representative results of more than 4 independent experiments.
Figure 3
Frequencies of DR0101 restricted PA401-420 reactive T cells and PA505-524 reactive T cells in DR0101 non-AVA vaccinees. A. PBMC from a DR0101 subject were stained with DR0101/PA401-420 PE-tetramers, DR0101/PA401-420 allophycocyanin -tetramers and gating antibodies ex vivo followed by enrichment with anti-PE magnetic beads. The frequency of PA401-420 specific T cells was determined to be 1.3 per million total CD4+ T cells. B. PBMC were stained with DR0101/PA401-420 PE-tetramers and CD45RA antibody, and gating antibodies ex vivo then enriched. The frequency of PA401-420 specific T cells was determined to be 0.5 per million total CD4+ T cells. C. PBMC from a DR0101 subject were stained with DR0101/PA505-524 PE-tetramers, DR0101/PA505-524 allophycocyanin-tetramers and gating antibodies ex vivo then enriched. The frequency of PA401-420 specific T cells was determined to be 0.2 per million total CD4+ T cells. Each plot represents staining from a different DR0101 subject. For each panel (A-C), we gated on forward and side scattering and then on cells that were Via-Probe-, CD4+ , CD14- and CD19-. Data shown in A and B are representative of more than 11 experiments in 11 subjects, and the data shown in C are representative of data from 7 different subjects.
Figure 4
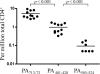
Frequencies of DR0101 restricted PA reactive T cells in DR0101 non-AVA vaccinees. Each symbol within each PA epitope represents staining for a different subject. Frequencies less than 0.1 per million were below the threshold of detection. The p values were determined by using two-tailed unpaired t tests.
Comment in
- Comment on "Frequency of epitope-specific naive CD4+ T cells correlates with immunodominance in the human memory repertoire".
Ascough S, Ingram RJ, Metan G, Maillere B, Doganay M, Ozkul Y, Kim LU, Ballie L, Moore S, Huwar TB, Sriskandan S, Altmann DM. Ascough S, et al. J Immunol. 2012 Jun 1;188(11):5205-6; author reply 5206. doi: 10.4049/jimmunol.1290018. J Immunol. 2012. PMID: 22611246 No abstract available.
Similar articles
- Comment on "Frequency of epitope-specific naive CD4+ T cells correlates with immunodominance in the human memory repertoire".
Ascough S, Ingram RJ, Metan G, Maillere B, Doganay M, Ozkul Y, Kim LU, Ballie L, Moore S, Huwar TB, Sriskandan S, Altmann DM. Ascough S, et al. J Immunol. 2012 Jun 1;188(11):5205-6; author reply 5206. doi: 10.4049/jimmunol.1290018. J Immunol. 2012. PMID: 22611246 No abstract available. - The anthrax vaccine adsorbed vaccine generates protective antigen (PA)-Specific CD4+ T cells with a phenotype distinct from that of naive PA T cells.
Kwok WW, Yang J, James E, Bui J, Huston L, Wiesen AR, Roti M. Kwok WW, et al. Infect Immun. 2008 Oct;76(10):4538-45. doi: 10.1128/IAI.00324-08. Epub 2008 Aug 4. Infect Immun. 2008. PMID: 18678674 Free PMC article. - Influence of antigenic experience on BJ gene segment usage in human CD4+ T cells.
Walser-Kuntz DR, Weyand CM, Goronzy JJ. Walser-Kuntz DR, et al. Int Immunol. 1997 Dec;9(12):1785-92. doi: 10.1093/intimm/9.12.1785. Int Immunol. 1997. PMID: 9466306 - Selection of immunodominant epitopes during antigen processing is hierarchical.
Sadegh-Nasseri S, Kim A. Sadegh-Nasseri S, et al. Mol Immunol. 2019 Sep;113:115-119. doi: 10.1016/j.molimm.2018.08.011. Epub 2018 Aug 24. Mol Immunol. 2019. PMID: 30146122 Free PMC article. Review. - CD4+ T cell epitope discovery and rational vaccine design.
Rosa DS, Ribeiro SP, Cunha-Neto E. Rosa DS, et al. Arch Immunol Ther Exp (Warsz). 2010 Apr;58(2):121-30. doi: 10.1007/s00005-010-0067-0. Epub 2010 Feb 14. Arch Immunol Ther Exp (Warsz). 2010. PMID: 20155490 Review.
Cited by
- T cell and antibody kinetics delineate SARS-CoV-2 peptides mediating long-term immune responses in COVID-19 convalescent individuals.
Bilich T, Nelde A, Heitmann JS, Maringer Y, Roerden M, Bauer J, Rieth J, Wacker M, Peter A, Hörber S, Rachfalski D, Märklin M, Stevanović S, Rammensee HG, Salih HR, Walz JS. Bilich T, et al. Sci Transl Med. 2021 Apr 21;13(590):eabf7517. doi: 10.1126/scitranslmed.abf7517. Epub 2021 Mar 15. Sci Transl Med. 2021. PMID: 33723016 Free PMC article. - Efficient ex vivo analysis of CD4+ T-cell responses using combinatorial HLA class II tetramer staining.
Uchtenhagen H, Rims C, Blahnik G, Chow IT, Kwok WW, Buckner JH, James EA. Uchtenhagen H, et al. Nat Commun. 2016 Aug 30;7:12614. doi: 10.1038/ncomms12614. Nat Commun. 2016. PMID: 27571776 Free PMC article. - Pegylation Reduces the Uptake of Certolizumab Pegol by Dendritic Cells and Epitope Presentation to T-Cells.
de Bourayne M, Meunier S, Bitoun S, Correia E, Mariette X, Nozach H, Maillère B. de Bourayne M, et al. Front Immunol. 2022 Feb 4;13:808606. doi: 10.3389/fimmu.2022.808606. eCollection 2022. Front Immunol. 2022. PMID: 35185895 Free PMC article. - Hybrid Insulin Peptides Are Recognized by Human T Cells in the Context of DRB1*04:01.
Arribas-Layton D, Guyer P, Delong T, Dang M, Chow IT, Speake C, Greenbaum CJ, Kwok WW, Baker RL, Haskins K, James EA. Arribas-Layton D, et al. Diabetes. 2020 Jul;69(7):1492-1502. doi: 10.2337/db19-0620. Epub 2020 Apr 14. Diabetes. 2020. PMID: 32291282 Free PMC article. - Analysis of Circulating Food Antigen-Specific T-Cells in Celiac Disease and Inflammatory Bowel Disease.
Rodríguez-Sillke Y, Schumann M, Lissner D, Branchi F, Proft F, Steinhoff U, Siegmund B, Glauben R. Rodríguez-Sillke Y, et al. Int J Mol Sci. 2023 May 2;24(9):8153. doi: 10.3390/ijms24098153. Int J Mol Sci. 2023. PMID: 37175860 Free PMC article.
References
- Whitmire JK, Benning N, Whitton JL. Precursor frequency, nonlinear proliferation, and functional maturation of virus-specific CD4+ T cells. J. Immunol. 2006;176:3028–3036. - PubMed
- McHeyzer-Williams MG, Davis MM. Antigen-specific development of primary and memory T cells in vivo. Science. 1995;268:106–111. - PubMed
- Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials