Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA - PubMed (original) (raw)
Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA
Liang Zhang et al. Nat Chem Biol. 2012.
Abstract
Human thymine DNA glycosylase (hTDG) efficiently excises 5-carboxylcytosine (5caC), a key oxidation product of 5-methylcytosine in genomic DNA, in a recently discovered cytosine demethylation pathway. We present here the crystal structures of the hTDG catalytic domain in complex with duplex DNA containing either 5caC or a fluorinated analog. These structures, together with biochemical and computational analyses, reveal that 5caC is specifically recognized in the active site of hTDG, supporting the role of TDG in mammalian 5-methylcytosine demethylation.
Conflict of interest statement
Competing financial interests
The authors declare no competing financial interests.
Figures
Figure 1. Electrophoretic mobility shift assay of hTDGcat(N140A) with 23mer dsDNA containing G•T, G•U, G•5fC and G•5caC pairs
(a) The proposed 5mC demethylation pathway. (b–c) The EMSA assay for hTDGcat(N140A) with dsDNA containing G•5fC and G•5caC pairs. The experiments were performed with 4 nM 32P-labeled DNA (40 pM 32P-labeled DNA for G•AP pairs) and various concentrations of hTDGcat(N140A) as shown.
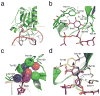
Figure 2. Schematic diagram of the hTDGcat(N140A)-A•5caC complex structure
(a) Overall structure of hTDGcat(N140A) bound with 5caC-containing dsDNA. The 5caC and the wedge residue Arg275 are shown as sticks and colored in magenta and green, respectively. (b) A network of hydrogen bonds in the active site of hTDG specifically recognizes 5caC. Residues involved in the interactions are labeled and shown as sticks, and hydrogen bonds are shown as yellow dashes. (c) The interactions involved in the 5-carboxyl-binding pocket. The atoms involved are presented as transparent spheres. (d) Superposition of 5caC and _β_-F-5caC in the active site pocket of the hTDGcat(N140A)-A•5caC and hTDGcat-G• _β_-F-5caC structures. Residues involved in the interactions in the _β_-F-5caC structure are shown as sticks and colored in yellow and orange. The fluorine atom is labeled in dark green. The hydrogen bond interaction between _β_-F-5caC and residue Ser271 is shown in yellow dashes.
Similar articles
- Structural Basis for Excision of 5-Formylcytosine by Thymine DNA Glycosylase.
Pidugu LS, Flowers JW, Coey CT, Pozharski E, Greenberg MM, Drohat AC. Pidugu LS, et al. Biochemistry. 2016 Nov 15;55(45):6205-6208. doi: 10.1021/acs.biochem.6b00982. Epub 2016 Nov 2. Biochemistry. 2016. PMID: 27805810 Free PMC article. - Excision of 5-hydroxymethyluracil and 5-carboxylcytosine by the thymine DNA glycosylase domain: its structural basis and implications for active DNA demethylation.
Hashimoto H, Hong S, Bhagwat AS, Zhang X, Cheng X. Hashimoto H, et al. Nucleic Acids Res. 2012 Nov 1;40(20):10203-14. doi: 10.1093/nar/gks845. Epub 2012 Sep 8. Nucleic Acids Res. 2012. PMID: 22962365 Free PMC article. - Activity and crystal structure of human thymine DNA glycosylase mutant N140A with 5-carboxylcytosine DNA at low pH.
Hashimoto H, Zhang X, Cheng X. Hashimoto H, et al. DNA Repair (Amst). 2013 Jul;12(7):535-40. doi: 10.1016/j.dnarep.2013.04.003. Epub 2013 May 13. DNA Repair (Amst). 2013. PMID: 23680598 Free PMC article. - Epigenetic modifications in DNA could mimic oxidative DNA damage: A double-edged sword.
Ito S, Kuraoka I. Ito S, et al. DNA Repair (Amst). 2015 Aug;32:52-57. doi: 10.1016/j.dnarep.2015.04.013. Epub 2015 May 1. DNA Repair (Amst). 2015. PMID: 25956859 Review. - Structural and mutation studies of two DNA demethylation related glycosylases: MBD4 and TDG.
Hashimoto H. Hashimoto H. Biophysics (Nagoya-shi). 2014 Oct 18;10:63-8. doi: 10.2142/biophysics.10.63. eCollection 2014. Biophysics (Nagoya-shi). 2014. PMID: 27493500 Free PMC article. Review.
Cited by
- TET proteins and the control of cytosine demethylation in cancer.
Scourzic L, Mouly E, Bernard OA. Scourzic L, et al. Genome Med. 2015 Jan 29;7(1):9. doi: 10.1186/s13073-015-0134-6. eCollection 2015. Genome Med. 2015. PMID: 25632305 Free PMC article. - Quantitative assessment of Tet-induced oxidation products of 5-methylcytosine in cellular and tissue DNA.
Liu S, Wang J, Su Y, Guerrero C, Zeng Y, Mitra D, Brooks PJ, Fisher DE, Song H, Wang Y. Liu S, et al. Nucleic Acids Res. 2013 Jul;41(13):6421-9. doi: 10.1093/nar/gkt360. Epub 2013 May 8. Nucleic Acids Res. 2013. PMID: 23658232 Free PMC article. - In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases.
Arand J, Spieler D, Karius T, Branco MR, Meilinger D, Meissner A, Jenuwein T, Xu G, Leonhardt H, Wolf V, Walter J. Arand J, et al. PLoS Genet. 2012 Jun;8(6):e1002750. doi: 10.1371/journal.pgen.1002750. Epub 2012 Jun 28. PLoS Genet. 2012. PMID: 22761581 Free PMC article. - TETonic shift: biological roles of TET proteins in DNA demethylation and transcription.
Pastor WA, Aravind L, Rao A. Pastor WA, et al. Nat Rev Mol Cell Biol. 2013 Jun;14(6):341-56. doi: 10.1038/nrm3589. Nat Rev Mol Cell Biol. 2013. PMID: 23698584 Free PMC article. Review. - A fungal dioxygenase CcTet serves as a eukaryotic 6mA demethylase on duplex DNA.
Mu Y, Zhang L, Hu J, Zhou J, Lin HW, He C, Chen HZ, Zhang L. Mu Y, et al. Nat Chem Biol. 2022 Jul;18(7):733-741. doi: 10.1038/s41589-022-01041-3. Epub 2022 Jun 2. Nat Chem Biol. 2022. PMID: 35654845
References
- Lindahl T. Nature. 1993;362:709–715. - PubMed
- Stivers JT, Jiang YL. Chem Rev. 2003;103:2729–2759. - PubMed
- Hitomi K, Iwai S, Tainer JA. DNA Repair (Amst) 2007;6:410–428. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases