PANDAseq: paired-end assembler for illumina sequences - PubMed (original) (raw)
PANDAseq: paired-end assembler for illumina sequences
Andre P Masella et al. BMC Bioinformatics. 2012.
Abstract
Background: Illumina paired-end reads are used to analyse microbial communities by targeting amplicons of the 16S rRNA gene. Publicly available tools are needed to assemble overlapping paired-end reads while correcting mismatches and uncalled bases; many errors could be corrected to obtain higher sequence yields using quality information.
Results: PANDAseq assembles paired-end reads rapidly and with the correction of most errors. Uncertain error corrections come from reads with many low-quality bases identified by upstream processing. Benchmarks were done using real error masks on simulated data, a pure source template, and a pooled template of genomic DNA from known organisms. PANDAseq assembled reads more rapidly and with reduced error incorporation compared to alternative methods.
Conclusions: PANDAseq rapidly assembles sequences and scales to billions of paired-end reads. Assembly of control libraries showed a 4-50% increase in the number of assembled sequences over naïve assembly with negligible loss of "good" sequence.
Figures
Figure 1
Schematic of paired-end assembly. Typical scenario: forward and reverse reads are overlapped and the primer regions are removed to reconstruct the sequences. Highly overlapping scenario: for short templates, the overlapping region may include the primer regions.
Figure 2
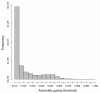
Quality scores of assembled masked data. A perfect 16S rRNA sequence from Sinorhizobium meliloti was masked using real Illumina quality scores and the resulting paired-end sequences were assembled with PANDAseq. A histogram of quality scores for the assembled sequences is shown.
Figure 3
Comparison of output of various assemblers. A scatter plot of the percentage of paired-end sequence assemblies from sequenced V3-region amplicons of Methylococcus capsulatus strain Bath against the average number of mismatching nucleotides between the assembled sequence and the reference sequence. The comparison was done between PANDAseq and three alternative assemblers (see text).
Figure 4
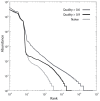
Rank abundance curves for control libraries. Rank-abundance curves for defined multi-organism libraries [1] assembled at two different quality thresholds using PANDAseq and naïve assembly followed by clustering with CD-HIT into OTUs of 97% identity.
References
- Bartram AK, Lynch MDJ, Stearns JC, Moreno-Hagelsieb G, Neufeld JD. Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end Illumina reads. Appl Environ Microbiol. 2011;77:3846–3852. doi: 10.1128/AEM.02772-10. http://aem.asm.org/cgi/content/abstract/77/11/3846 - DOI - PMC - PubMed
- Degnan PH, Ochman H. Illumina-based analysis of microbial community diversity. ISME J. 2011. http://www.nature.com/ismej/journal/v6/n1/full/ismej201174a.html - PMC - PubMed
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4516–4522. http://genomebiology.com/2011/12/5/R50 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases