Structural basis for specific recognition of Rpt1p, an ATPase subunit of 26 S proteasome, by proteasome-dedicated chaperone Hsm3p - PubMed (original) (raw)
Structural basis for specific recognition of Rpt1p, an ATPase subunit of 26 S proteasome, by proteasome-dedicated chaperone Hsm3p
Kenji Takagi et al. J Biol Chem. 2012.
Abstract
The 26 S proteasome is a 2.5-MDa molecular machine that degrades ubiquitinated proteins in eukaryotic cells. It consists of a proteolytic core particle and two 19 S regulatory particles (RPs) composed of 6 ATPase (Rpt) and 13 non-ATPase (Rpn) subunits. Multiple proteasome-dedicated chaperones facilitate the assembly of the proteasome, but little is known about the detailed mechanisms. Hsm3, a 19 S RP dedicated chaperone, transiently binds to the C-terminal domain of the Rpt1 subunit and forms a tetrameric complex, Hsm3-Rpt1-Rpt2-Rpn1, during maturation of the ATPase ring of 19 S RP. To elucidate the structural basis of Hsm3 function, we determined the crystal structures of Hsm3 and its complex with the C-terminal domain of the Rpt1 subunit (Rpt1C). Hsm3 has a C-shaped structure that consists of 11 HEAT repeats. The structure of the Hsm3-Rpt1C complex revealed that the interacting surface between Hsm3 and Rpt1 is a hydrophobic core and a complementary charged surface. Mutations in the Hsm3-Rpt1 surface resulted in the assembly defect of the 26 S proteasome. Furthermore, a structural model of the Hsm3-Rpt ring complex and an in vitro binding assay suggest that Hsm3 can bind Rpt2 in addition to Rpt1. Collectively, our results provide the structural basis of the molecular functions of Hsm3 for the RP assembly.
Figures
FIGURE 1.
Three-dimensional structure of yeast Hsm3. A, Two orthogonal views of the crystal structure of Hsm3. Each HEAT repeat of Hsm3 is labeled with HR1–HR11. The N and C termini of molecule are indicated. B, topological diagram of the secondary structure elements of Hsm3. The α-helices are represented by red circles (helix A in HR) and blue circles (helix B in HR), and the 310-helix is represented by yellow circles. Superscripts on the circles indicate the number of start and end residues of HEAT repeats. C, ribbon diagram of the two Hsm3 monomers in the asymmetric unit of the crystal. The N and C termini of each molecule are indicated.
FIGURE 2.
Structure of the Hsm3 complexed with the C-terminal domain of Rpt1. A, overall architecture of the Hsm3-Rpt1C complex. Hsm3 (chain A) and Rpt1C (chain B) are colored green and orange, respectively. The N and C termini of each molecule are indicated. B, ribbon diagram of Rpt1C structure. Four α-helices are labeled from α1 to α4. C, structural comparison of C-terminal domains of three AAA+-ATPases as follows: Rpt1C (orange), PAN (PDB code 3H4M, cyan), and HslU (PDB code 1DO0, green). D, structural comparison between the free and the Rpt1C-bound Hsm3. The free Hsm3 (green) and the Hsm3 (red) in the complex with Rpt1C (orange) are shown.
FIGURE 3.
Interface between Hsm3 and Rpt1. A, close-up view of the Hsm3-Rpt1C interface. A ribbon representation of the interacting regions of the complex. Some of the potential interacting residues are shown by stick representation. Hsm3 and Rpt1C are colored green and orange, respectively. B, surface potential representation of the Hsm3 and Rpt1C. The complementary surface patches responsible for complex formation are depicted by a circle. Red, blue, and white represent acidic, basic, and neutral, respectively.
FIGURE 4.
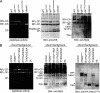
Characterization of the crucial residues in the Hsm3-Rpt1 interface. A, pulldown assay from bacterially co-expressing His6-Hsm3 and mutant versions of Rpt1C. Eluted proteins from Ni-NTA resin were subjected to SDS-PAGE followed by Coomassie Brilliant Blue (CBB) staining. B, pulldown assay from bacterially co-expressing His6-Hsm3 mutants and Rpt1C. Eluted proteins from Ni-NTA resin were subjected to SDS-PAGE followed by Coomassie Brilliant Blue staining. Triple mutation of Hsm3 (E157A/T190A/D230A) is indicated in Fig. 3_A. C,_ pulldown assay between full-length His6-Rpt1 and Hsm3 mutants. His6-fused full-length Rpt1 and indicated Hsm3 mutants were co-synthesized by a wheat germ cell-free expression system and then pulled down by Ni-NTA resin. Input and eluted proteins were subjected to SDS-PAGE followed by Coomassie Brilliant Blue staining. Total reaction and supernatant are labeled as T and S, respectively.
FIGURE 5.
Mutations of the Hsm3-Rpt1 interface cause assembly defect of the 26 S proteasome. A, native-PAGE analysis of the 26 S proteasome assembly in rpt1 mutants. Extracts of the indicated mutants were subjected to native-PAGE. After electrophoresis, peptidase activities of the proteasome were visualized by in gel peptidase assay (left). The same gel was analyzed by Western blotting with the indicated antibodies. RP_2_-CP and RP_1_-CP indicate doubly and singly capped 26 S proteasomes, respectively. Rpn1 and Rpt2 are abbreviated as N1 and T2, respectively. B, native-PAGE analysis of the 26 S proteasome assembly in hsm3 mutants. HA-tagged Hsm3 and its mutants were expressed under the native promoter in the Δ_hsm3_ cells. Extracts from the indicated mutants were analyzed as in A. RP* and base* seemed to be transient complexes that contain Hsm3. Hsm3 mutants that harbor triple mutations of Hsm3 (E157A/T190A/D230A) are indicated as 3A.
FIGURE 6.
Hsm3 is a scaffold protein for Rpt1-Rpt2 formation. A, structural model of Hsm3-Rpt ring complex. The Hsm3-Rpt1C structure was superimposed on a structural model of hexameric ATPase (ATPase ring model was generated from the HslU (PDB code 1DO0) structure). Hsm3 and Rpt ring are shown as Cα trace representations and are colored green (Hsm3), orange (Rpt1), cyan (Rpt2), blue (Rpt3), red (Rpt4), purple (Rpt5), and pink (Rpt6), respectively. Surfaces of Rpt subunits are shown as surface plots (gray). B, in vitro pulldown assay between Hsm3 and His6-tagged base subunits. His6-tagged full-length Rpt1, Rpt2, Rpt5, or Rpn1 were synthesized with or without Hsm3 by a wheat germ cell-free expression system. Interactions between the His6-tagged base subunits and Hsm3 were analyzed by pulldown assay with Ni-NTA resin. Input and eluted proteins were subjected to SDS-PAGE followed by Coomassie Brilliant Blue (CBB) staining. The protein bands corresponding to the base subunits and Hsm3 are indicated by filled and open arrowheads, respectively. C, model of the base assembly. Four assembly intermediates are formed from nine free base subunits and a deubiquitylating enzyme Ubp6 (42). This study suggests that Hsm3 plays a scaffolding role in the formation of the Rpt1 and Rpt2 complex. Finally, the four intermediates are jointed into the base subcomplex. Nas2 is dissociated upon binding of Nas2 module and Nas6 module (43). Rpn and Rpt are abbreviated as N and T, respectively.
Similar articles
- Dual functions of the Hsm3 protein in chaperoning and scaffolding regulatory particle subunits during the proteasome assembly.
Barrault MB, Richet N, Godard C, Murciano B, Le Tallec B, Rousseau E, Legrand P, Charbonnier JB, Le Du MH, Guérois R, Ochsenbein F, Peyroche A. Barrault MB, et al. Proc Natl Acad Sci U S A. 2012 Apr 24;109(17):E1001-10. doi: 10.1073/pnas.1116538109. Epub 2012 Mar 29. Proc Natl Acad Sci U S A. 2012. PMID: 22460800 Free PMC article. - Hexameric assembly of the proteasomal ATPases is templated through their C termini.
Park S, Roelofs J, Kim W, Robert J, Schmidt M, Gygi SP, Finley D. Park S, et al. Nature. 2009 Jun 11;459(7248):866-70. doi: 10.1038/nature08065. Nature. 2009. PMID: 19412160 Free PMC article. - Crystal structure of yeast rpn14, a chaperone of the 19 S regulatory particle of the proteasome.
Kim S, Saeki Y, Fukunaga K, Suzuki A, Takagi K, Yamane T, Tanaka K, Mizushima T, Kato K. Kim S, et al. J Biol Chem. 2010 May 14;285(20):15159-15166. doi: 10.1074/jbc.M110.104042. Epub 2010 Mar 16. J Biol Chem. 2010. PMID: 20236927 Free PMC article. - Assembly manual for the proteasome regulatory particle: the first draft.
Park S, Tian G, Roelofs J, Finley D. Park S, et al. Biochem Soc Trans. 2010 Feb;38(Pt 1):6-13. doi: 10.1042/BST0380006. Biochem Soc Trans. 2010. PMID: 20074027 Free PMC article. Review. - Proteasome activator 200: the heat is on..
Savulescu AF, Glickman MH. Savulescu AF, et al. Mol Cell Proteomics. 2011 May;10(5):R110.006890. doi: 10.1074/mcp.R110.006890. Epub 2011 Mar 9. Mol Cell Proteomics. 2011. PMID: 21389348 Free PMC article. Review.
Cited by
- Structure, Dynamics and Function of the 26S Proteasome.
Mao Y. Mao Y. Subcell Biochem. 2021;96:1-151. doi: 10.1007/978-3-030-58971-4_1. Subcell Biochem. 2021. PMID: 33252727 Review. - Reconfiguration of the proteasome during chaperone-mediated assembly.
Park S, Li X, Kim HM, Singh CR, Tian G, Hoyt MA, Lovell S, Battaile KP, Zolkiewski M, Coffino P, Roelofs J, Cheng Y, Finley D. Park S, et al. Nature. 2013 May 23;497(7450):512-6. doi: 10.1038/nature12123. Epub 2013 May 5. Nature. 2013. PMID: 23644457 Free PMC article. - Effect of cryogenic grinding on volatile and fatty oil constituents of cumin (Cuminum cyminum L.) genotypes.
Sharma LK, Agarwal D, Rathore SS, Malhotra SK, Saxena SN. Sharma LK, et al. J Food Sci Technol. 2016 Jun;53(6):2827-34. doi: 10.1007/s13197-016-2258-0. Epub 2016 May 27. J Food Sci Technol. 2016. PMID: 27478239 Free PMC article. - Molecular architecture and assembly of the eukaryotic proteasome.
Tomko RJ Jr, Hochstrasser M. Tomko RJ Jr, et al. Annu Rev Biochem. 2013;82:415-45. doi: 10.1146/annurev-biochem-060410-150257. Epub 2013 Mar 13. Annu Rev Biochem. 2013. PMID: 23495936 Free PMC article. Review. - Dynamic Regulation of the 26S Proteasome: From Synthesis to Degradation.
Marshall RS, Vierstra RD. Marshall RS, et al. Front Mol Biosci. 2019 Jun 7;6:40. doi: 10.3389/fmolb.2019.00040. eCollection 2019. Front Mol Biosci. 2019. PMID: 31231659 Free PMC article. Review.
References
- Baumeister W., Walz J., Zühl F., Seemüller E. (1998) The proteasome. Paradigm of a self-compartmentalizing protease. Cell 92, 367–380 - PubMed
- Coux O., Tanaka K., Goldberg A. L. (1996) Structure and functions of the 20 S and 26 S proteasomes. Annu. Rev. Biochem. 65, 801–847 - PubMed
- Glickman M. H., Rubin D. M., Coux O., Wefes I., Pfeifer G., Cjeka Z., Baumeister W., Fried V. A., Finley D. (1998) A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94, 615–623 - PubMed
- Leggett D. S., Glickman M. H., Finley D. (2005) Purification of proteasomes, proteasome subcomplexes, and proteasome-associated proteins from budding yeast. Methods Mol. Biol. 301, 57–70 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous