Opposing functions of the ETS factor family define Shh spatial expression in limb buds and underlie polydactyly - PubMed (original) (raw)
. 2012 Feb 14;22(2):459-67.
doi: 10.1016/j.devcel.2011.12.010.
Iain Williamson, John H Wiltshire, Silvia Peluso, Paul S Devenney, Alison E Hill, Abdelkader Essafi, James Hagman, Richard Mort, Graeme Grimes, Carlo L DeAngelis, Robert E Hill
Affiliations
- PMID: 22340503
- PMCID: PMC3314984
- DOI: 10.1016/j.devcel.2011.12.010
Opposing functions of the ETS factor family define Shh spatial expression in limb buds and underlie polydactyly
Laura A Lettice et al. Dev Cell. 2012.
Abstract
Sonic hedgehog (Shh) expression during limb development is crucial for specifying the identity and number of digits. The spatial pattern of Shh expression is restricted to a region called the zone of polarizing activity (ZPA), and this expression is controlled from a long distance by the cis-regulator ZRS. Here, members of two groups of ETS transcription factors are shown to act directly at the ZRS mediating a differential effect on Shh, defining its spatial expression pattern. Occupancy at multiple GABPα/ETS1 sites regulates the position of the ZPA boundary, whereas ETV4/ETV5 binding restricts expression outside the ZPA. The ETS gene family is therefore attributed with specifying the boundaries of the classical ZPA. Two point mutations within the ZRS change the profile of ETS binding and activate Shh expression at an ectopic site in the limb bud. These molecular changes define a pathogenetic mechanism that leads to preaxial polydactyly (PPD).
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
Graphical abstract
Figure 1
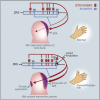
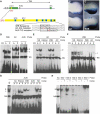
Point Mutations Alter Shh Expression and Protein Binding Profiles (A) Schematic showing the ZRS (yellow box), which resides within intron 5 of LMBR1, 1 Mb for the Shh gene. The positions of the ETS sites 1–5 and ETV sites A and B identified within the ZRS are marked by blue and green boxes, respectively. The sequences around the mutations identified in families with PPD (Family AC and AUS) are shown. (B and C) Limbs from transgenic animals carrying wild-type (B) and mutant (C) ZRS reporter constructs (forelimb buds are shown on top and hindlimb buds below) demonstrate that the AUS mutation results in expansion of the posterior expression (compare to B) and ectopic staining in the anterior mesenchyme (arrows). (D–H) EMSA analysis of nuclear extracts from anterior (A) and posterior (P) halves of E11.5 limb buds. (D) Nuclear extract was incubated with ds-oligos containing the WT, AC, or AUS sequence. The WT sequence produced a specific band (1); the AC point mutation resulted in a higher migrating band (2); and the AUS mutation produced a combination of WT and AC binding; bands 1 and 2. A nonspecific (NS) band was observed for all ds-oligos. (E–G) EMSA using the AC ds-oligo (E), WT ds-oligo (F), and AUS ds-oligo (G), and using an unlabeled NS sequence, ETS consensus sequence (EtsCon), WT, or AC oligonucleotide as their competitors. (H) Comparison by EMSA of the binding for the wild-type ZRS sites 1–5, showing a greater extent of binding to the AC mutant site and sites 1 and 3. The unlabelled AC oligonucleotide (lanes labeled +) specifically competes for band 2.
Figure 2
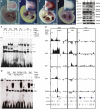
ETS Factors Are Expressed in the Limb and Bind to the ZRS (A–D) Whole-mount in situ hybridization analysis for Ets1 (A), Ets2 (B), Etv4 (C), and Etv5 (D) are shown in E11.5 embryos and limb buds. (E) Western blot analysis using antibodies raised against ETS factors, designated αETV4, αETV5, αETS2, αELF1, αETS1, and αGABPα, and against histone H3 (αH3) and actin (αactin), with nuclear extracts from the anterior (A) and posterior (P) halves of the limb buds (E11.5). Also shown is a comparison between limb nuclear extracts (NE) and cytoplasmic extracts (CE). αH3 and αactin were used as loading controls. (F) EMSA shows WtB and AC ds-oligo binding in nuclear extracts depleted for ETV4 or ETS1 using specific antibodies (αEtv4 and αEts1). (IgG was used as a nonspecific control.) Extracts from anterior (A) or posterior (P) halves or whole limbs (L) from E11.5 limb buds were used. Band 1 observed with the WtB probe was specifically depleted by the addition of αETV4 antibody, while Band shift 2 observed with the AC probe was specifically depleted by the addition of αETS1 antibody. (G) EMSAs were conducted with ds-oligos containing the sequence for the wild-type ETV4 site A (WtA) or the Belg2 mutation (Bg). WtA ds-oligo shows a specific band (1) while that for Bg sequence gives an additional higher migrating band (2). The anti-ETV4 antibody depletes Band 1 observed with WtA and Bg probes (nonspecific IgG used as control). (H) ChIP using antibodies to five different ETS factors (ETV4, ETV5, Elf1, ETS1, and GABPα) analyzed by hybridizing to tiling microarrays. Summary is presented using three different genomic regions, the y axis is Log2 for each ChIP/input DNA and the x axis represents a segment of DNA from the microarray. The DNA region containing the ZRS is highlighted by the gray shading. As controls, the whole of the Shh coding region plus promoter (Shh) and the region downstream of the α-globin locus (3′Hba) are shown. Scale bars are shown at the top and the positions of potential ETS1/GABPα (AGGAAG/A) and ETV4/ETV5 (AGAAAG/A) binding sites at the bottom of each panel.
Figure 3
Transgenic Analysis of Embryos Carrying Mutant ZRS Sequences (A–D) Limbs from transgenic embryos carrying the following mutant ZRS sequences: the 44 bp terminal deletion (tDel) (A), the 3 bp change in the ETV4 Site B (-ETVB) (B), and Site A (-ETVA) (C). Disruption of both sites in combination (D) results in ectopic expression in the anterior of the limb. (E–G) Position of the run of Ts within the ZRS and the changes added are shown in red. Expression due to these changes is shown by comparison of the addition of seven Ts (F) and the extra ETS1/GABPα site (AGGAAGT) (G). Ectopic expression is detected in (G). (H) Graphical representation of the expression pattern resulting from mutations within the endogenous ETS sites. Expression pattern is the ratio of the width of the expression domain divided by the width of the limb, expressed as a percentage (see p values in Table S2). (I–Q) Examples of transgenic embryos for the ETS mutations analyzed are shown. The ETS sites remaining are depicted in the lower-left-hand corner of each figure. (R–T) Transgenic embryos that represent the addition of the AUS mutation in combination with ETS site mutations are shown. A close-up of a forelimb for each is shown in the insets.
Figure 4
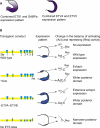
A Model Representing the Fine Balance of ETS Factor Binding and Their Effects on Shh Expression (A) Representation of the expression patterns of the activating ETS factors (ETS1 and GABPα) and the repressing ETS factors (ETV4 and ETV5). (B) A summary diagram of how four of the transgenic constructs are proposed to interact with the available ETS factors in the limb, with the expression pattern observed for each construct shown in the middle. The change in the balance between activating and repressing activity represented on the right shows the relative balance in the anterior and posterior margins of the limb. The size of the lettering represents the relative amounts of the activating and repressing activities.
Similar articles
- FGF-regulated Etv genes are essential for repressing Shh expression in mouse limb buds.
Zhang Z, Verheyden JM, Hassell JA, Sun X. Zhang Z, et al. Dev Cell. 2009 Apr;16(4):607-13. doi: 10.1016/j.devcel.2009.02.008. Dev Cell. 2009. PMID: 19386269 Free PMC article. - Etv2 regulates enhancer chromatin status to initiate Shh expression in the limb bud.
Koyano-Nakagawa N, Gong W, Das S, Theisen JWM, Swanholm TB, Van Ly D, Dsouza N, Singh BN, Kawakami H, Young S, Chen KQ, Kawakami Y, Garry DJ. Koyano-Nakagawa N, et al. Nat Commun. 2022 Jul 21;13(1):4221. doi: 10.1038/s41467-022-31848-6. Nat Commun. 2022. PMID: 35864091 Free PMC article. - How to make a zone of polarizing activity: insights into limb development via the abnormality preaxial polydactyly.
Hill RE. Hill RE. Dev Growth Differ. 2007 Aug;49(6):439-48. doi: 10.1111/j.1440-169X.2007.00943.x. Dev Growth Differ. 2007. PMID: 17661738 Review. - How is digit identity determined during limb development?
Suzuki T. Suzuki T. Dev Growth Differ. 2013 Jan;55(1):130-8. doi: 10.1111/dgd.12022. Epub 2012 Dec 12. Dev Growth Differ. 2013. PMID: 23230964 Review.
Cited by
- Mutagenesis Sensitivity Mapping of Human Enhancers In Vivo.
Kosicki M, Zhang B, Pampari A, Akiyama JA, Plajzer-Frick I, Novak CS, Tran S, Zhu Y, Kato M, Hunter RD, von Maydell K, Barton S, Beckman E, Kundaje A, Dickel DE, Visel A, Pennacchio LA. Kosicki M, et al. bioRxiv [Preprint]. 2024 Sep 8:2024.09.06.611737. doi: 10.1101/2024.09.06.611737. bioRxiv. 2024. PMID: 39282388 Free PMC article. Preprint. - DNA-binding factor footprints and enhancer RNAs identify functional non-coding genetic variants.
Biddie SC, Weykopf G, Hird EF, Friman ET, Bickmore WA. Biddie SC, et al. Genome Biol. 2024 Aug 6;25(1):208. doi: 10.1186/s13059-024-03352-1. Genome Biol. 2024. PMID: 39107801 Free PMC article. - TBX3 is essential for establishment of the posterior boundary of anterior genes and upregulation of posterior genes together with HAND2 during the onset of limb bud development.
Soussi G, Girdziusaite A, Jhanwar S, Palacio V, Notaro M, Sheth R, Zeller R, Zuniga A. Soussi G, et al. Development. 2024 Jun 1;151(11):dev202722. doi: 10.1242/dev.202722. Epub 2024 Jun 3. Development. 2024. PMID: 38828908 Free PMC article. - Conserved _Cis_-Acting Range Extender Element Mediates Extreme Long-Range Enhancer Activity in Mammals.
Bower G, Hollingsworth EW, Jacinto S, Clock B, Cao K, Liu M, Dziulko A, Alcaina-Caro A, Xu Q, Skowronska-Krawczyk D, Lopez-Rios J, Dickel DE, Bardet AF, Pennacchio LA, Visel A, Kvon EZ. Bower G, et al. bioRxiv [Preprint]. 2024 May 26:2024.05.26.595809. doi: 10.1101/2024.05.26.595809. bioRxiv. 2024. PMID: 38826394 Free PMC article. Preprint.
References
- Ahn S., Joyner A.L. Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell. 2004;118:505–516. - PubMed
- Amano T., Sagai T., Tanabe H., Mizushina Y., Nakazawa H., Shiroishi T. Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Dev. Cell. 2009;16:47–57. - PubMed
- Efron B. Correlation and large-scale simultaneous significance testing. J. Am. Stat. Assoc. 2007;102:93–103.
- Farooq M., Troelsen J.T., Boyd M., Eiberg H., Hansen L., Hussain M.S., Rehman S., Azhar A., Ali A., Bakhtiar S.M. Preaxial polydactyly/triphalangeal thumb is associated with changed transcription factor-binding affinity in a family with a novel point mutation in the long-range cis-regulatory element ZRS. Eur. J. Hum. Genet. 2010;18:733–736. - PMC - PubMed
- Fisher R.J., Koizumi S., Kondoh A., Mariano J.M., Mavrothalassitis G., Bhat N.K., Papas T.S. Human ETS1 oncoprotein. Purification, isoforms, -SH modification, and DNA sequence-specific binding. J. Biol. Chem. 1992;267:17957–17965. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous