Meta-analysis of untargeted metabolomic data from multiple profiling experiments - PubMed (original) (raw)
Meta-analysis of untargeted metabolomic data from multiple profiling experiments
Gary J Patti et al. Nat Protoc. 2012.
Abstract
metaXCMS is a software program for the analysis of liquid chromatography/mass spectrometry-based untargeted metabolomic data. It is designed to identify the differences between metabolic profiles across multiple sample groups (e.g., 'healthy' versus 'active disease' versus 'inactive disease'). Although performing pairwise comparisons alone can provide physiologically relevant data, these experiments often result in hundreds of differences, and comparison with additional biologically meaningful sample groups can allow for substantial data reduction. By performing second-order (meta-) analysis, metaXCMS facilitates the prioritization of interesting metabolite features from large untargeted metabolomic data sets before the rate-limiting step of structural identification. Here we provide a detailed step-by-step protocol for going from raw mass spectrometry data to metaXCMS results, visualized as Venn diagrams and exported Microsoft Excel spreadsheets. There is no upper limit to the number of sample groups or individual samples that can be compared with the software, and data from most commercial mass spectrometers are supported. The speed of the analysis depends on computational resources and data volume, but will generally be less than 1 d for most users. metaXCMS is freely available at http://metlin.scripps.edu/metaxcms/.
Figures
Figure 1
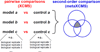
Introduction of pairwise and second-order comparison. XCMS performs a pairwise comparison of two sample groups with any number of biological replicates. Data from multiple pairwise comparisons is then used by metaXCMS to perform a second-order comparison in which shared and unique differences are identified.
Figure 2
Data reduction by meta-analysis. Three pairwise comparisons of different pain models to their respective controls resulted in 22,577 detected metabolite features (model A is animals plantar injected with Complete Freund’s Adjuvant, model B is animals treated with noxious heat, and model C is animals intraperitoneally injected with serum from K/BxN mice, for further details see Tautenhahn et al.). Next, features with fold changes less than 1.5 and _p_-values greater than 0.05 were filtered and the remaining 1,825 features were plotted. A second-order comparison by metaXCMS showed that only 3 of these features were commonly shared, one of which was determined to be histamine.
Figure 3
Visualization of theoretical meta-analysis applied to identify biomarkers of disease severity. The left Venn diagram shows shared and unique metabolite features for mild disease, severe disease, and healthy patients. While features in the areas labeled a, b, and c may serve as biomarkers, areas a and c could provide additional markers specific to mild and severe disease respectively. The right Venn diagram shows a second-order visualization of the same comparison that is representative of metaXCMS output when the parameters are set to plot only metabolite features that are unique to disease (i.e., features that are detected in disease but not in healthy samples). The advantage of the second-order visualization is that it is not limited to representing only metabolites unique to a certain sample group. Rather, metabolites that are up- and down-regulated by even small fold changes can be easily represented according to user-defined thresholds. Given that biomarkers may not be metabolites unique to disease samples but instead metabolites that increase by some quantified fold change, second-order visualizations are generally better suited for metabolomic data since they can be used to show up- and down-regulated features (see Venn diagram in Figure 2). Changing the second-order visualization here to include features with smaller fold changes, for example, would result in the display of more features that might represent useful diagnostic markers.
Figure 4
Overview of the computational workflow. The workflow consists of five stages: acquisition of LC/MS data, conversion of the data to .mzXML files, analysis of the files by XCMS, analysis of XCMS results by metaXCMS, and result browsing and interpretation.
Figure 5
MSConvertGUI.exe, the graphical user interface of the ProteoWizard file converter. The input fields or icons of the software that correspond to specific steps in the protocol are indicated by the step number.
Figure 6
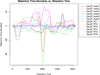
Retention-time correction curves generated by XCMS. Each colored line represents a different sample processed. Note that the retention-time deviation is different for each sample and that it is not linear.
Figure 7
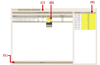
Graphical user interface of metaXCMS. The input fields or icons related to the import of XCMS diffreports are indicated by arrows that are numbered according to the protocol step in which they are described.
Figure 8
Graphical user interface of metaXCMS. Filtering may be performed on the basis of _p_-value, fold change, and up-/down-regulation. The input fields or icons related to filtering are indicated by arrows that are numbered according to the protocol step in which they are described.
Figure 9
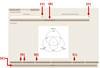
Graphical user interface of metaXCMS. Features that are uniquely or commonly altered among the pairwise comparisons are displayed as Venn diagrams. The icons related to data visualization and export are indicated by arrows that are numbered according to the protocol step in which they are described.
Figure 10
Graphical user interface of metaXCMS. Retention-time correction for all samples compared is displayed and EICs are generated. The icons related to retention-time correction and EIC generation are indicated by arrows that are numbered according to the protocol step in which they are described.
Similar articles
- metaXCMS: second-order analysis of untargeted metabolomics data.
Tautenhahn R, Patti GJ, Kalisiak E, Miyamoto T, Schmidt M, Lo FY, McBee J, Baliga NS, Siuzdak G. Tautenhahn R, et al. Anal Chem. 2011 Feb 1;83(3):696-700. doi: 10.1021/ac102980g. Epub 2010 Dec 21. Anal Chem. 2011. PMID: 21174458 Free PMC article. - Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the METLIN database.
Zhu ZJ, Schultz AW, Wang J, Johnson CH, Yannone SM, Patti GJ, Siuzdak G. Zhu ZJ, et al. Nat Protoc. 2013 Mar;8(3):451-60. doi: 10.1038/nprot.2013.004. Epub 2013 Feb 7. Nat Protoc. 2013. PMID: 23391889 Free PMC article. - Translational Metabolomics of Head Injury: Exploring Dysfunctional Cerebral Metabolism with Ex Vivo NMR Spectroscopy-Based Metabolite Quantification.
Wolahan SM, Hirt D, Glenn TC. Wolahan SM, et al. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. PMID: 26269925 Free Books & Documents. Review. - After the feature presentation: technologies bridging untargeted metabolomics and biology.
Cho K, Mahieu NG, Johnson SL, Patti GJ. Cho K, et al. Curr Opin Biotechnol. 2014 Aug;28:143-8. doi: 10.1016/j.copbio.2014.04.006. Epub 2014 May 6. Curr Opin Biotechnol. 2014. PMID: 24816495 Free PMC article. Review. - Autonomous metabolomics for rapid metabolite identification in global profiling.
Benton HP, Ivanisevic J, Mahieu NG, Kurczy ME, Johnson CH, Franco L, Rinehart D, Valentine E, Gowda H, Ubhi BK, Tautenhahn R, Gieschen A, Fields MW, Patti GJ, Siuzdak G. Benton HP, et al. Anal Chem. 2015 Jan 20;87(2):884-91. doi: 10.1021/ac5025649. Epub 2014 Dec 26. Anal Chem. 2015. PMID: 25496351 Free PMC article.
Cited by
- Tracing the Biotransformation of Polycyclic Aromatic Hydrocarbons in Contaminated Soil Using Stable Isotope-Assisted Metabolomics.
Tian Z, Vila J, Yu M, Bodnar W, Aitken MD. Tian Z, et al. Environ Sci Technol Lett. 2018 Feb 13;5(2):103-109. doi: 10.1021/acs.estlett.7b00554. Epub 2018 Jan 2. Environ Sci Technol Lett. 2018. PMID: 31572742 Free PMC article. - PAIRUP-MS: Pathway analysis and imputation to relate unknowns in profiles from mass spectrometry-based metabolite data.
Hsu YH, Churchhouse C, Pers TH, Mercader JM, Metspalu A, Fischer K, Fortney K, Morgen EK, Gonzalez C, Gonzalez ME, Esko T, Hirschhorn JN. Hsu YH, et al. PLoS Comput Biol. 2019 Jan 14;15(1):e1006734. doi: 10.1371/journal.pcbi.1006734. eCollection 2019 Jan. PLoS Comput Biol. 2019. PMID: 30640898 Free PMC article. - Integrating untargeted metabolomics, genetically informed causal inference, and pathway enrichment to define the obesity metabolome.
Hsu YH, Astley CM, Cole JB, Vedantam S, Mercader JM, Metspalu A, Fischer K, Fortney K, Morgen EK, Gonzalez C, Gonzalez ME, Esko T, Hirschhorn JN. Hsu YH, et al. Int J Obes (Lond). 2020 Jul;44(7):1596-1606. doi: 10.1038/s41366-020-0603-x. Epub 2020 May 28. Int J Obes (Lond). 2020. PMID: 32467615 Free PMC article. - Engineered microfluidic bioreactor for examining the three-dimensional breast tumor microenvironment.
Rogers M, Sobolik T, Schaffer DK, Samson PC, Johnson AC, Owens P, Codreanu SG, Sherrod SD, McLean JA, Wikswo JP, Richmond A. Rogers M, et al. Biomicrofluidics. 2018 May 7;12(3):034102. doi: 10.1063/1.5016433. eCollection 2018 May. Biomicrofluidics. 2018. PMID: 29774083 Free PMC article. - Metabolic regulator LKB1 is crucial for Schwann cell-mediated axon maintenance.
Beirowski B, Babetto E, Golden JP, Chen YJ, Yang K, Gross RW, Patti GJ, Milbrandt J. Beirowski B, et al. Nat Neurosci. 2014 Oct;17(10):1351-61. doi: 10.1038/nn.3809. Epub 2014 Sep 7. Nat Neurosci. 2014. PMID: 25195104 Free PMC article.
References
- Baker M. Metabolomics: from small molecules to big ideas. Nat Meth. 2011;8:117–121.
Publication types
MeSH terms
Substances
Grants and funding
- P01 DA026146-02/DA/NIDA NIH HHS/United States
- R24 EY017540-04/EY/NEI NIH HHS/United States
- L30 AG0 038036/AG/NIA NIH HHS/United States
- P30 MH062261/MH/NIMH NIH HHS/United States
- P01 DA026146/DA/NIDA NIH HHS/United States
- L30 AG038036/AG/NIA NIH HHS/United States
- P30 MH062261-10/MH/NIMH NIH HHS/United States
- R01 CA170737/CA/NCI NIH HHS/United States
- R24 EY017540/EY/NEI NIH HHS/United States
LinkOut - more resources
Full Text Sources