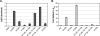Mechanism of substrate selection by a highly specific CRISPR endoribonuclease - PubMed (original) (raw)
Mechanism of substrate selection by a highly specific CRISPR endoribonuclease
Samuel H Sternberg et al. RNA. 2012 Apr.
Abstract
Bacteria and archaea possess adaptive immune systems that rely on small RNAs for defense against invasive genetic elements. CRISPR (clustered regularly interspaced short palindromic repeats) genomic loci are transcribed as long precursor RNAs, which must be enzymatically cleaved to generate mature CRISPR-derived RNAs (crRNAs) that serve as guides for foreign nucleic acid targeting and degradation. This processing occurs within the repetitive sequence and is catalyzed by a dedicated Cas6 family member in many CRISPR systems. In Pseudomonas aeruginosa, crRNA biogenesis requires the endoribonuclease Csy4 (Cas6f), which binds and cleaves at the 3' side of a stable RNA stem-loop structure encoded by the CRISPR repeat. We show here that Csy4 recognizes its RNA substrate with an ~50 pM equilibrium dissociation constant, making it one of the highest-affinity protein:RNA interactions of this size reported to date. Tight binding is mediated exclusively by interactions upstream of the scissile phosphate that allow Csy4 to remain bound to its product and thereby sequester the crRNA for downstream targeting. Substrate specificity is achieved by RNA major groove contacts that are highly sensitive to helical geometry, as well as a strict preference for guanosine adjacent to the scissile phosphate in the active site. Collectively, our data highlight diverse modes of substrate recognition employed by Csy4 to enable accurate selection of CRISPR transcripts while avoiding spurious, off-target RNA binding and cleavage.
Figures
FIGURE 1.
Csy4 binds its substrate and product with high affinity and functions as a single-turnover enzyme. (A) Csy4 cleaves within pre-crRNA repeat sequences (black) to generate mature crRNAs that contain a spacer sequence (colored line) flanked by fragments of the repeat. The substrate sequence and cleavage site (red triangle) are indicated above, with the crRNA construct previously used for crystallography shown in bold. (B) A schematic depicts protein:RNA contacts revealed by a co-crystal structure of Csy4 bound to a fragment of the crRNA repeat (PDB ID: 2XLK). Important amino acid residues are shown in yellow, and RNA nucleotides are numbered as in A. Red circles, pentagons, boxes, and red dotted lines denote phosphates, ribose groups, bases, and hydrogen-bonding interactions, respectively. (C) EMSAs (top) were performed with Csy4-H29A and the substrate and product of the cleavage reaction. The resulting data for these and all subsequent binding assays were fit with a standard binding isotherm to yield equilibrium dissociation constants (solid lines; see Materials and Methods), and average _K_d and standard error of the mean (SEM) values from at least three independent experiments are reported in Supplemental Table 1. (D) RNA cleavage assays were conducted at five different enzyme:substrate molar ratios, and the extent of the reaction at various time points was assessed by denaturing PAGE (top). The resulting data for these and all subsequent cleavage assays were fit with a single exponential to yield first-order rate constants (solid lines; see Materials and Methods), and average kobs and SEM values from three independent experiments are reported in Supplemental Table 1. Error bars for each time point represent the standard deviation and are not always visible.
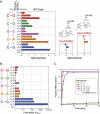
FIGURE 2.
Amino acid contributions to binding energy and cleavage kinetics. (A) Csy4 residues involved in base-pair recognition and phosphate backbone contacts were mutated to alanine in order to assess their energetic contributions to binding. EMSAs were performed with a noncleavable crRNA repeat substrate containing a deoxyribonucleotide substitution at G20, and binding defects relative to Csy4-H29A were determined and converted to ΔΔG values (T = 298 K). Plotted are the average and SEM from at least three independent experiments. (B) First-order rate constants (kobs) for WT-crRNA repeat cleavage by each Csy4 mutant were determined. Cleavage data for R118A/R115A and R115A/R119A mutants showed biphasic kinetics and were fit with a double exponential decay to yield two rate constants (Supplemental Table 2), the faster of which is shown. Plotted are the average fold defects (relative to WT-Csy4) and SEM from three independent experiments. Average _K_d, kobs, and SEM values are reported in Supplemental Table 2.
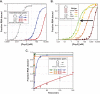
FIGURE 3.
Importance of loop sequence for high-affinity RNA binding. (A) EMSAs demonstrate that Csy4 binds the reverse complement of the crRNA repeat (rc) >105-fold more weakly than the WT-crRNA repeat. (B) Mutant RNA substrates were generated by changing the WT loop sequence (GUAUA) to a quintuple mutant (UAUAC), the highly stable UUCG tetraloop, or a poly(A) pentaloop, or by removing the loop through use of a substrate nicked between U12 and A13. EMSAs reveal substantial defects associated with binding these mutant RNAs.
FIGURE 4.
Substrate specificity within the crRNA repeat stem. (A) A library of mutated crRNA repeat substrates was generated containing all possible Watson-Crick base-pair substitutions at each position in the double-stranded stem. EMSAs were performed with Csy4-H29A and these RNA substrates, and the resulting binding defects relative to WT-crRNA repeat were determined and converted to ΔΔG values (T = 298 K). The WT stem sequence is shown at the left, with data for base-pair substitutions at each position color-coded similarly. Binding experiments with RNAs mutated at the bottom C–G or U–A base pair were repeated with Csy4-R102A (middle) or Csy4-Q104A (right), respectively; ΔΔG values were calculated relative to WT-crRNA repeat binding by each Csy4 mutant. Shown above are chemical structures of the interactions made by Arg102 and Gln104 with the WT base pairs. (B) Single-turnover cleavage assays were performed with the same library of RNA mutants as in A, and the resulting defects in kobs relative to WT-crRNA repeat were determined. The data are plotted as in A. (C) To investigate the importance of the terminal C–G base pair during cleavage, mismatched substrates were generated by mutating C6 or G20 individually and single-turnover cleavage assays were performed.
FIGURE 5.
Stem length dependence during substrate binding and cleavage. (A) One or two G–C base pairs were inserted at the top of the stem between the closing C–G base pair and the GUAUA pentaloop, and EMSAs were performed. (B) To test the hypothesis that longer stems prevent stable binding of the arginine-rich helix via their effect on major groove accessibility, a substrate was generated that contains five G–C base pairs inserted above the WT stem. Subsequently, asymmetric adenosine bulges were inserted on the 3′ side of the duplex between the five-base-pair WT stem and the five-base-pair insertion. EMSAs reveal that binding affinities increase monotonically (black arrow) with bulges of increasing size. (C) One or two G–C or A–U base pairs were inserted below the terminal C–G base pair, and cleavage time courses were performed. Additional A–U base pairs have negligible effects on kobs, whereas two additional G–C base pairs result in ∼1500-fold slower kinetics.
Similar articles
- Sequence- and structure-specific RNA processing by a CRISPR endonuclease.
Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Haurwitz RE, et al. Science. 2010 Sep 10;329(5997):1355-8. doi: 10.1126/science.1192272. Science. 2010. PMID: 20829488 Free PMC article. - Csy4 relies on an unusual catalytic dyad to position and cleave CRISPR RNA.
Haurwitz RE, Sternberg SH, Doudna JA. Haurwitz RE, et al. EMBO J. 2012 Jun 13;31(12):2824-32. doi: 10.1038/emboj.2012.107. Epub 2012 Apr 20. EMBO J. 2012. PMID: 22522703 Free PMC article. - Cutting it close: CRISPR-associated endoribonuclease structure and function.
Hochstrasser ML, Doudna JA. Hochstrasser ML, et al. Trends Biochem Sci. 2015 Jan;40(1):58-66. doi: 10.1016/j.tibs.2014.10.007. Epub 2014 Nov 18. Trends Biochem Sci. 2015. PMID: 25468820 Review. - Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity.
Charpentier E, Richter H, van der Oost J, White MF. Charpentier E, et al. FEMS Microbiol Rev. 2015 May;39(3):428-41. doi: 10.1093/femsre/fuv023. Epub 2015 May 19. FEMS Microbiol Rev. 2015. PMID: 25994611 Free PMC article. Review. - Binding and cleavage of CRISPR RNA by Cas6.
Carte J, Pfister NT, Compton MM, Terns RM, Terns MP. Carte J, et al. RNA. 2010 Nov;16(11):2181-8. doi: 10.1261/rna.2230110. Epub 2010 Sep 30. RNA. 2010. PMID: 20884784 Free PMC article.
Cited by
- crRNA and tracrRNA guide Cas9-mediated DNA interference in Streptococcus thermophilus.
Karvelis T, Gasiunas G, Miksys A, Barrangou R, Horvath P, Siksnys V. Karvelis T, et al. RNA Biol. 2013 May;10(5):841-51. doi: 10.4161/rna.24203. Epub 2013 Mar 27. RNA Biol. 2013. PMID: 23535272 Free PMC article. - Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection.
East-Seletsky A, O'Connell MR, Knight SC, Burstein D, Cate JH, Tjian R, Doudna JA. East-Seletsky A, et al. Nature. 2016 Oct 13;538(7624):270-273. doi: 10.1038/nature19802. Epub 2016 Sep 26. Nature. 2016. PMID: 27669025 Free PMC article. - Characterization of CRISPR RNA biogenesis and Cas6 cleavage-mediated inhibition of a provirus in the haloarchaeon Haloferax mediterranei.
Li M, Liu H, Han J, Liu J, Wang R, Zhao D, Zhou J, Xiang H. Li M, et al. J Bacteriol. 2013 Feb;195(4):867-75. doi: 10.1128/JB.01688-12. Epub 2012 Dec 14. J Bacteriol. 2013. PMID: 23243301 Free PMC article. - Dynamic modulation of enzyme activity by synthetic CRISPR-Cas6 endonucleases.
Mitkas AA, Valverde M, Chen W. Mitkas AA, et al. Nat Chem Biol. 2022 May;18(5):492-500. doi: 10.1038/s41589-022-01005-7. Epub 2022 Apr 25. Nat Chem Biol. 2022. PMID: 35468950 - Primed adaptation tolerates extensive structural and size variations of the CRISPR RNA guide in Haloarcula hispanica.
Gong L, Li M, Cheng F, Zhao D, Chen Y, Xiang H. Gong L, et al. Nucleic Acids Res. 2019 Jun 20;47(11):5880-5891. doi: 10.1093/nar/gkz244. Nucleic Acids Res. 2019. PMID: 30957847 Free PMC article.
References
- Al-Attar S, Westra ER, van der Oost J, Brouns SJJ 2011. Clustered regularly interspaced short palindromic repeats (CRISPRs): The hallmark of an ingenious antiviral defense mechanism in prokaryotes. Biol Chem 392: 277–289 - PubMed
- Busso D, Delagoutte-Busso B, Moras D 2005. Construction of a set Gateway-based destination vectors for high-throughput cloning and expression screening in Escherichia coli. Anal Biochem 343: 313–321 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources