A 17q12 allele is associated with altered NK cell subsets and function - PubMed (original) (raw)
A 17q12 allele is associated with altered NK cell subsets and function
Zongqi Xia et al. J Immunol. 2012.
Abstract
NK cells play an important role in innate immunity. A previous genome-wide association study demonstrated an association between a 17q12 allele (rs9916629(C)) and lower frequency of CD3(-)CD56(+) NK cells in peripheral blood. We performed an analysis that not only replicates the original result of the genome-wide association study (p = 0.036) but also defines the specific cell subpopulations and functions that are modulated by the rs9916629 polymorphism in a cohort of 96 healthy adult subjects using targeted multiparameter flow cytometric profiling of NK cell phenotypes and functions. We found that rs9916629(C) is associated with alterations in specific NK cell subsets, including lower frequency of predominantly cytotoxic CD56(dim) NK cells (p = 0.011), higher frequency of predominantly regulatory CD56(bright) NK cells (p = 0.019), and a higher proportion of NK cells expressing the inhibitory NKG2A receptor (p = 0.0002). Functionally, rs9916629(C) is associated with decreased secretion of macrophage inflammatory protein-1β by NK cells in the context of Ab-dependent cell-mediated cytotoxicity (p = 0.039) and increased degranulation in response to MHC class I-deficient B cells (p = 0.017). Transcriptional profiling of NK cells suggests that rs9916629 influences the expression of transcription factors such as TBX21, which has a role in NK cell differentiation, offering a possible mechanism for the phenotypic and functional differences between the different alleles. The rs9916629(C) allele therefore has a validated effect on the proportion of NK cells in peripheral blood and skews NK cells toward a specific phenotypic and functional profile, potentially influencing the impact that these innate immune cells have on infection and autoimmunity.
Conflict of interest statement
Disclosures
The authors have no financial conflicts of interest.
Figures
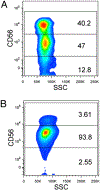
FIGURE 1.
Representative flow cytometry data for the primary outcome from two subjects. (A) Homozygote for the reference allele C; (B) homozygote for the T allele at the rs9916629 SNP.
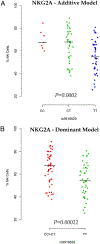
FIGURE 2.
The effect of rs9916629C on the frequency of NKG2A-expressing NK subpopulation using additive (A) and dominant model (B). We compare the homozygotes for the alternative allele (TT) to the combined heterozygotes (CT) and homozygotes (CC) for the reference C allele. The reference allele frequency is 0.31.
Similar articles
- Distinct natural killer cells in HIV-exposed seronegative subjects with effector cytotoxic CD56(dim) and CD56(bright) cells and memory-like CD57⁺NKG2C⁺CD56(dim) cells.
Lima JF, Oliveira LM, Pereira NZ, Mitsunari GE, Duarte AJ, Sato MN. Lima JF, et al. J Acquir Immune Defic Syndr. 2014 Dec 15;67(5):463-71. doi: 10.1097/QAI.0000000000000350. J Acquir Immune Defic Syndr. 2014. PMID: 25230289 - Specific phenotype and function of CD56-expressing innate immune cell subsets in human thymus.
Gerstner S, Köhler W, Heidkamp G, Purbojo A, Uchida S, Ekici AB, Heger L, Luetke-Eversloh M, Schubert R, Bader P, Klingebiel T, Koehl U, Mackensen A, Romagnani C, Cesnjevar R, Dudziak D, Ullrich E. Gerstner S, et al. J Leukoc Biol. 2016 Dec;100(6):1297-1310. doi: 10.1189/jlb.1A0116-038R. Epub 2016 Jun 28. J Leukoc Biol. 2016. PMID: 27354408 - Selective depletion of CD56(dim) NK cell subsets and maintenance of CD56(bright) NK cells in treatment-naive HIV-1-seropositive individuals.
Tarazona R, Casado JG, Delarosa O, Torre-Cisneros J, Villanueva JL, Sanchez B, Galiani MD, Gonzalez R, Solana R, Peña J. Tarazona R, et al. J Clin Immunol. 2002 May;22(3):176-83. doi: 10.1023/a:1015476114409. J Clin Immunol. 2002. PMID: 12078859 - The biology of human natural killer-cell subsets.
Cooper MA, Fehniger TA, Caligiuri MA. Cooper MA, et al. Trends Immunol. 2001 Nov;22(11):633-40. doi: 10.1016/s1471-4906(01)02060-9. Trends Immunol. 2001. PMID: 11698225 Review. - The Broad Spectrum of Human Natural Killer Cell Diversity.
Freud AG, Mundy-Bosse BL, Yu J, Caligiuri MA. Freud AG, et al. Immunity. 2017 Nov 21;47(5):820-833. doi: 10.1016/j.immuni.2017.10.008. Immunity. 2017. PMID: 29166586 Free PMC article. Review.
Cited by
- The natural killer cell-associated rs9916629-C allele is a novel genetic risk factor for fatal COVID-19.
Vietzen H, Furlano PL, Traugott M, Totschnig D, Hoepler W, Strassl R, Zoufaly A, Puchhammer-Stöckl E. Vietzen H, et al. J Med Virol. 2023 Jan;95(1):e28404. doi: 10.1002/jmv.28404. J Med Virol. 2023. PMID: 36515427 Free PMC article. - Clinical relevance and functional consequences of the TNFRSF1A multiple sclerosis locus.
Ottoboni L, Frohlich IY, Lee M, Healy BC, Keenan BT, Xia Z, Chitnis T, Guttmann CR, Khoury SJ, Weiner HL, Hafler DA, De Jager PL. Ottoboni L, et al. Neurology. 2013 Nov 26;81(22):1891-9. doi: 10.1212/01.wnl.0000436612.66328.8a. Epub 2013 Oct 30. Neurology. 2013. PMID: 24174586 Free PMC article. - Landscape of X chromosome inactivation across human tissues.
Tukiainen T, Villani AC, Yen A, Rivas MA, Marshall JL, Satija R, Aguirre M, Gauthier L, Fleharty M, Kirby A, Cummings BB, Castel SE, Karczewski KJ, Aguet F, Byrnes A; GTEx Consortium; Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group; Statistical Methods groups—Analysis Working Group; Enhancing GTEx (eGTEx) groups; NIH Common Fund; NIH/NCI; NIH/NHGRI; NIH/NIMH; NIH/NIDA; Biospecimen Collection Source Site—NDRI; Biospecimen Collection Source Site—RPCI; Biospecimen Core Resource—VARI; Brain Bank Repository—University of Miami Brain Endowment Bank; Leidos Biomedical—Project Management; ELSI Study; Genome Browser Data Integration &Visualization—EBI; Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz; Lappalainen T, Regev A, Ardlie KG, Hacohen N, MacArthur DG. Tukiainen T, et al. Nature. 2017 Oct 11;550(7675):244-248. doi: 10.1038/nature24265. Nature. 2017. PMID: 29022598 Free PMC article. - HLA-C levels impact natural killer cell subset distribution and function.
Sips M, Liu Q, Draghi M, Ghebremichael M, Berger CT, Suscovich TJ, Sun Y, Walker BD, Carrington M, Altfeld M, Brouckaert P, De Jager PL, Alter G. Sips M, et al. Hum Immunol. 2016 Dec;77(12):1147-1153. doi: 10.1016/j.humimm.2016.08.004. Epub 2016 Aug 10. Hum Immunol. 2016. PMID: 27521484 Free PMC article. - Coordinating GWAS results with gene expression in a systems immunologic paradigm in autoimmunity.
Stranger BE, De Jager PL. Stranger BE, et al. Curr Opin Immunol. 2012 Oct;24(5):544-51. doi: 10.1016/j.coi.2012.09.002. Epub 2012 Oct 3. Curr Opin Immunol. 2012. PMID: 23040211 Free PMC article. Review.
References
- Berger CT, and Alter G. 2011. Natural killer cells in spontaneous control of HIV infection. Curr. Opin. HIV AIDS 6: 208–213. - PubMed
- Cerwenka A, and Lanier LL. 2001. Natural killer cells, viruses and cancer. Nat. Rev. Immunol 1: 41–49. - PubMed
- Cooper MA, Fehniger TA, and Caligiuri MA. 2001. The biology of human natural killer-cell subsets. Trends Immunol. 22: 633–640. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous