Kruppel-like factor 4 (KLF4) promotes the survival of natural killer cells and maintains the number of conventional dendritic cells in the spleen - PubMed (original) (raw)
Kruppel-like factor 4 (KLF4) promotes the survival of natural killer cells and maintains the number of conventional dendritic cells in the spleen
Chun Shik Park et al. J Leukoc Biol. 2012 May.
Abstract
The development and survival of NK cells rely on a complex, spatiotemporal gene expression pattern regulated by specific transcription factors in NK cells and tissue-specific microenvironments supported by hematopoietic cells. Here, we show that somatic deletion of the KLF4 gene, using inducible and lineage-specific cre-transgenic mice, leads to a significant reduction of NK cells (NK1.1(+) TCR-β(-)) in the blood and spleen but not in the BM, liver, or LNs. Functional and immunophenotypic analyses revealed increased apoptosis of CD27(+/-) CD11b(+) NK cells in the spleen of KLF4-deficient mice, although remaining NK cells were able to lyse tumor target cells and produce IFN-γ. A normal recovery of adoptively transferred KLF4-deficient NK cells in WT hosts suggested that the survival defect was not intrinsic of NK cells. However, BM chimeras using KLF4-deficient mice as donors indicated that reduced survival of NK cells depended on BM-derived hematopoietic cells in the spleen. The number of CD11c(hi) DCs, which are known to support NK cell survival, was reduced significantly in the spleen of KLF4-deficient mice, likely a result of a lower number of precDC progenitor cells in this tissue. Taken together, our data suggest that the pluripotency-associated gene KLF4 is required for the maintenance of DCs in the spleen and consequently, survival of differentiated NK cells in this tissue.
Figures
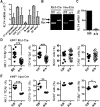
Figure 1.. Loss of KLF4 function leads to a reduction of NK cells in peripheral blood.
(A) KLF4 transcript levels were measured by real-time quantitative PCR in granulocytes (CD11b+ Gr1+), Mo (CD11b+ Gr1−), CD8+ T cells, CD4+ T cells, B cells (B220+), NK cells (NK1.1+ TCR-β−), and DCs (CD11chi; _n_=3). (B) Deletion of the Klf4 gene was confirmed by PCR using genomic DNA from BM cells isolated from _Klf4_fl/fl _Mx1_-Cre− (fl/fl) and _Klf4_fl/fl _Mx1_-Cre+ (Δ/Δ) mice, 3 months after poly (I:C) administration, and _Klf4_fl/fl _Vav_-iCre+ (Δ/Δ) and _Klf4_fl/fl _Vav_-iCre− mice (fl/fl). (C) KLF4 expression was measured by real-time quantitative PCR in purified NK cells from _Klf4_fl/fl _Mx1_-Cre+ (Δ/Δ) and _Klf4_fl/fl _Mx1_-Cre− (fl/fl) mice, 3 months after gene deletion. (D) Percentages of NK cells, Mo, B cells, and T cells were measured in blood 5 months after poly (I:C) administration in _Klf4_fl/fl _Mx1_-Cre− (fl/fl) and _Klf4_fl/fl _Mx1_-Cre+ (Δ/Δ) mice (_n_=12). (E) Determination of NK cells, Mo, B cells, and T cells in the blood of 3-month-old _Klf4_fl/fl _Vav_-iCre+ (Δ/Δ) and _Klf4_fl/fl _Vav_-iCre− mice (fl/fl). Each symbol represents an individual mouse (_n_=5). *P < 0.05; **P < 0.005; ***P < 0.0005 (unpaired Student's t test).
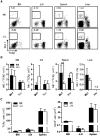
Figure 2.. KLF4 regulates homeostatic maintenance of NK cells in the spleen.
(A) Flow cytometric analysis of NK1.1+ TCR-β− NK cells in the BM, iLNs, spleen, and liver of _Klf4_fl/fl _Mx1_-Cre+ (Δ/Δ) and _Klf4_fl/fl _Mx1_-Cre− (fl/fl) mice. Numbers represent the percentage of cells in an outlined gate of representative dot plots. (B) Total numbers (BM, iLN, spleen) and percentages (liver) of NK cells were determined in _Klf4_fl/fl carrying the _Mx1_-Cre (Mx1) or _Vav_-iCre (Vav) transgenes (fl/fl, closed bars; Δ/Δ, open bars; _n_=8). n.d., Not determined. (C) Total numbers of T cells (TCR-β+; left panel) and B cells (B220+; right panel) in BM, iLN, and the spleen of _Klf4_fl/fl _Vav_-iCre− (fl/fl, closed bars) and _Klf4_fl/fl _Vav_-iCre+ (Δ/Δ, open bars) mice (_n_=6). **P < 0.005 (unpaired Student's t test).
Figure 3.. mNK cell subsets are reduced in blood and the spleen of KLF4-deficient mice.
(A) Flow cytometric analysis of NK cell subsets based on the expression of CD27 and CD11b in NK1.1+ TCR-β− cells: CD27+ CD11b− (Subset I), CD27+ CD11b+ (Subset II), and CD27− CD11b+ (Subset III). A representative dot plot and distribution are shown for BM, liver, blood, and the spleen of _Klf4_fl/fl _Mx1_-Cre+ (Δ/Δ) and _Klf4_fl/fl _Mx1_-Cre− (fl/fl) mice (_n_=6). (B) Flow cytometric analysis of NKP (CD11b− CD122+ TCR-β− NK1.1−), iNK (CD11b− CD122+ TCR-β− NK1.1+), and mNK (CD122+ NK1.1+ TCR-β− CD11b+) cells. Total numbers of NKP, iNK, and mNK cells are shown in the BM of _Klf4_fl/fl _Vav_-iCre− (fl/fl, closed bars) and _Klf4_fl/fl _Vav_-iCre+ (Δ/Δ, open bars) mice (_n_=8). *P < 0.05; **P < 0.005; ***P < 0.0005 (unpaired Student's t test).
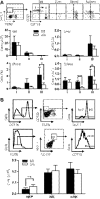
Figure 4.. Increased apoptosis in KLF4-deficient mNK cells.
(A) Apoptosis of NK Subsets I–III in the spleen was determined by Annexin V staining in _Klf4_fl/fl _Vav_-iCre+ (Δ/Δ, open bars) and _Klf4_fl/fl _Vav_-iCre− (fl/fl, closed bars) mice. Representative data are shown on the left and statistical analysis on the right (_n_=6). FSC, Forward-scatter. (B) Intracellular detection of proapoptotic proteins Bim and Noxa in CD11b− and CD11b+ NK cells isolated from the spleen of _Klf4_fl/fl _Vav_-iCre− (fl/fl, dotted lines) and _Klf4_fl/fl _Vav_-iCre+ (Δ/Δ, solid lines) mice. (C) Statistical analysis of Bim and Noxa in CD11b− and CD11b+ NK cells from the _Klf4_fl/fl _Vav_-iCre− (fl/fl, closed circles) and _Klf4_fl/fl _Vav_-iCre+ (Δ/Δ, open circles) mice (_n_=8). MFI, Mean fluorescence intensity. *P < 0.05; **P < 0.005; ***P < 0.0005 (unpaired Student's t test).
Figure 5.. BM-derived hematopoietic cells support survival of NK cells in the spleen of KLF4-deficient mice.
(A) NK cells were purified from the spleen of _Klf4_fl/fl _Vav_-iCre− (fl/fl) and _Klf4_fl/fl _Vav_-iCre+ (Δ/Δ) mice (CD45.2+) and adoptively transferred into nonirradiated WT mice (B6.SJL, CD45.1+) to study NK cell dependency on impaired survival. Numbers of transferred NK cells recovered in the spleen and BM after 10 days are shown on the right. (B) To study the role of KLF4 on radio-resistant stromal cells, BM cells from WT mice (B6.SJL, CD45.1+) were transplanted into lethally irradiated _Klf4_fl/fl _Mx1_-Cre− (fl/fl) or _Klf4_fl/fl _Mx1_-Cre+ (Δ/Δ) mice (both CD45.2+). The numbers of donor-derived NK cells (CD45.1+ NK1.1+ TCR-β−) were determined in blood, BM, and the spleen, 2 months after transplant (_n_=4–6). (C) BM cells from _Klf4_fl/fl _Vav_-iCre− (fl/fl) or _Klf4_fl/fl _Vav_-iCre+ (Δ/Δ) mice (CD45.2+) were transplanted into lethally irradiated WT mice (B6.SJL, CD45.1+) to examine the role of hematopoietic cells in NK cell survival (_n_=4–5). Three months after transplantation, the contribution of donor-derived NK cells (CD45.2+ NK1.1+ TCR-β−) was monitored in the spleen, blood, and BM by flow cytometry. *P <0.05 (unpaired Student's t test).
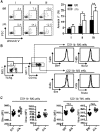
Figure 6.. KLF4 regulates the pool of cDCs in the spleen.
(A) Representative flow cytometric determination of cDC (CD11chi CD11b+), Mo (CD11clo CD11bint), and Mφ (CD11clo CD11bhi; left panel). Total numbers of cDCs, Mo, and Mφ in the spleen of _Klf4_fl/fl _Vav_-iCre− (fl/fl) and _Klf4_fl/fl _Vav_-iCre+ (Δ/Δ) mice are shown on the right (_n_=8–9). (B) Annexin V staining of cDCs, Mo, and Mφ isolated from the spleen of _Klf4_fl/fl _Vav_-iCre− (fl/fl, closed circles) or _Klf4_fl/fl _Vav_-iCre+ (Δ/Δ, open circles) mice (_n_=8–9). (C) Numbers of precDCs (CD11cint CD45RAlo CD43int SIRPαint CD11b− CD4− CD8− cells) were determined in the spleen of _Klf4_fl/fl _Vav_-iCre− (fl/fl, closed circles) or _Klf4_fl/fl _Vav_-iCre+ (Δ/Δ, open circles) mice by flow cytometry (_n_=8–9). (D) Percentage of donor-derived cDCs in the spleen of WT recipient mice (B6.SJL), transplanted with _Klf4_fl/fl _Vav_-iCre− (fl/fl) or _Klf4_fl/fl _Vav_-iCre+ (Δ/Δ) BM cells (_n_=5). (E) KLF4 expression in cDCs isolated from the spleen of _Klf4_fl/fl _Vav_-iCre− (fl/fl) or _Klf4_fl/fl _Vav_-iCre+ (Δ/Δ) mice (_n_=3). (F) Transcript levels of IL-15 in cDCs isolated from the spleen of _Klf4_fl/fl _Vav_-iCre− (fl/fl) or _Klf4_fl/fl _Vav_-iCre+ (Δ/Δ) mice (_n_=3). **P < 0.005; ***P < 0.0005 (unpaired Student's t test).
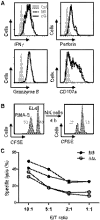
Figure 7.. KLF4 is dispensable for the expression of effector molecules and cytotoxicity in NK cells.
(A) Intracellular levels of IFN-γ, granzyme B, and perforin and cell-surface expression of CD107a were analyzed in _Klf4_fl/fl _Mx1_-Cre− (fl/fl, dotted lines) and _Klf4_fl/fl _Mx1_-Cre+ (Δ/Δ, solid lines) NK cells by flow cytometry. The gray, filled histograms correspond to an unstained control sample. (B) Representative flow cytometric analysis of the in vitro NK cell cytolytic assay using a mixture of CFSE-labeled RMA-S cells (targets) and EL4 cells (control), incubated NK1.1+ cells purified from the spleen of _Klf4_fl/fl _Mx1_-Cre+ (Δ/Δ) and _Klf4_fl/fl _Mx1_-Cre− (fl/fl) mice. (C) Specific lysis calculated based on the percentage of CFSElow (RMA-S) and CFSEhigh (EL4) cells at different E:T ratios. Data are representative of two independent experiments (two mice/genotype).
Similar articles
- Impaired NK cell development in an IFN-gamma transgenic mouse: aberrantly expressed IFN-gamma enhances hematopoietic stem cell apoptosis and affects NK cell differentiation.
Shimozato O, Ortaldo JR, Komschlies KL, Young HA. Shimozato O, et al. J Immunol. 2002 Feb 15;168(4):1746-52. doi: 10.4049/jimmunol.168.4.1746. J Immunol. 2002. PMID: 11823506 - IFN regulatory factor-2 deficiency revealed a novel checkpoint critical for the generation of peripheral NK cells.
Taki S, Nakajima S, Ichikawa E, Saito T, Hida S. Taki S, et al. J Immunol. 2005 May 15;174(10):6005-12. doi: 10.4049/jimmunol.174.10.6005. J Immunol. 2005. PMID: 15879093 - Natural killer dendritic cells have both antigen presenting and lytic function and in response to CpG produce IFN-gamma via autocrine IL-12.
Pillarisetty VG, Katz SC, Bleier JI, Shah AB, Dematteo RP. Pillarisetty VG, et al. J Immunol. 2005 Mar 1;174(5):2612-8. doi: 10.4049/jimmunol.174.5.2612. J Immunol. 2005. PMID: 15728467 - Monocytes control natural killer cell differentiation to effector phenotypes.
Soderquest K, Powell N, Luci C, van Rooijen N, Hidalgo A, Geissmann F, Walzer T, Lord GM, Martín-Fontecha A. Soderquest K, et al. Blood. 2011 Apr 28;117(17):4511-8. doi: 10.1182/blood-2010-10-312264. Epub 2011 Mar 9. Blood. 2011. PMID: 21389319 - The transcription factor E4BP4 is not required for extramedullary pathways of NK cell development.
Crotta S, Gkioka A, Male V, Duarte JH, Davidson S, Nisoli I, Brady HJ, Wack A. Crotta S, et al. J Immunol. 2014 Mar 15;192(6):2677-88. doi: 10.4049/jimmunol.1302765. Epub 2014 Feb 17. J Immunol. 2014. PMID: 24534532 Free PMC article.
Cited by
- Foxm1 haploinsufficiency drives clonal hematopoiesis and promotes a stress-related transition to hematologic malignancy in mice.
Yu C, Sheng Y, Yu F, Ni H, Qiu A, Huang Y, Qian Z. Yu C, et al. J Clin Invest. 2023 Aug 1;133(15):e163911. doi: 10.1172/JCI163911. J Clin Invest. 2023. PMID: 37526082 Free PMC article. - Krüppel-like Factor 4 Supports the Expansion of Leukemia Stem Cells in MLL-AF9-driven Acute Myeloid Leukemia.
Lewis AH, Bridges CS, Moorshead DN, Chen TJ, Du W, Zorman B, Sumazin P, Puppi M, Lacorazza HD. Lewis AH, et al. Stem Cells. 2022 Aug 25;40(8):736-750. doi: 10.1093/stmcls/sxac033. Stem Cells. 2022. PMID: 35535819 Free PMC article. - Identification and Targeting of the Developmental Blockade in Extranodal Natural Killer/T-cell Lymphoma.
Mundy-Bosse BL, Weigel C, Wu YZ, Abdelbaky S, Youssef Y, Casas SB, Polley N, Ernst G, Young KA, McConnell KK, Nalin AP, Wu KG, Broughton M, Lordo MR, Altynova E, Hegewisch-Solloa E, Enriquez-Vera DY, Dueñas D, Barrionuevo C, Yu SC, Saleem A, Suarez CJ, Briercheck EL, Molina-Kirsch H, Loughran TP, Weichenhan D, Plass C, Reneau JC, Mace EM, Gamboa FV, Weinstock DM, Natkunam Y, Caligiuri MA, Mishra A, Porcu P, Baiocchi RA, Brammer JE, Freud AG, Oakes CC. Mundy-Bosse BL, et al. Blood Cancer Discov. 2022 Mar 1;3(2):154-169. doi: 10.1158/2643-3230.BCD-21-0098. Blood Cancer Discov. 2022. PMID: 35247900 Free PMC article. - Krüppel-like factor 4 promotes survival and expansion in acute myeloid leukemia cells.
Lewis AH, Bridges CS, Punia VS, Cooper AFJ, Puppi M, Lacorazza HD. Lewis AH, et al. Oncotarget. 2021 Feb 16;12(4):255-267. doi: 10.18632/oncotarget.27878. eCollection 2021 Feb 16. Oncotarget. 2021. PMID: 33659038 Free PMC article. - Recent Discoveries on the Involvement of Krüppel-Like Factor 4 in the Most Common Cancer Types.
Taracha-Wisniewska A, Kotarba G, Dworkin S, Wilanowski T. Taracha-Wisniewska A, et al. Int J Mol Sci. 2020 Nov 22;21(22):8843. doi: 10.3390/ijms21228843. Int J Mol Sci. 2020. PMID: 33266506 Free PMC article. Review.
References
- Vosshenrich C. A., Garcia-Ojeda M. E., Samson-Villeger S. I., Pasqualetto V., Enault L., Richard-Le Goff O., Corcuff E., Guy-Grand D., Rocha B., Cumano A., Rogge L., Ezine S., Di Santo J. P. (2006) A thymic pathway of mouse natural killer cell development characterized by expression of GATA-3 and CD127. Nat. Immunol. 7, 1217–1224 - PubMed
- Freud A. G., Becknell B., Roychowdhury S., Mao H. C., Ferketich A. K., Nuovo G. J., Hughes T. L., Marburger T. B., Sung J., Baiocchi R. A., Guimond M., Caligiuri M. A. (2005) A human CD34(+) subset resides in lymph nodes and differentiates into CD56bright natural killer cells. Immunity 22, 295–304 - PubMed
- Lanier L. L. (2005) NK cell recognition. Annu. Rev. Immunol. 23, 225–274 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K12 GM084897/GM/NIGMS NIH HHS/United States
- R01 AI077536/AI/NIAID NIH HHS/United States
- R01AI077536/AI/NIAID NIH HHS/United States
- R01AI077536-02S1/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials