Bacterial biogeography of the human digestive tract - PubMed (original) (raw)
Bacterial biogeography of the human digestive tract
Jennifer C Stearns et al. Sci Rep. 2011.
Abstract
We present bacterial biogeography as sampled from the human gastrointestinal tract of four healthy subjects. This study generated >32 million paired-end sequences of bacterial 16S rRNA genes (V3 region) representing >95,000 unique operational taxonomic units (OTUs; 97% similarity clusters), with >99% Good's coverage for all samples. The highest OTU richness and phylogenetic diversity was found in the mouth samples. The microbial communities of multiple biopsy sites within the colon were highly similar within individuals and largely distinct from those in stool. Within an individual, OTU overlap among broad site definitions (mouth, stomach/duodenum, colon and stool) ranged from 32-110 OTUs, 25 of which were common to all individuals and included OTUs affiliated with Faecalibacterium prasnitzii and the TM7 phylum. This first comprehensive characterization of the abundant and rare microflora found along the healthy human digestive tract represents essential groundwork to investigate further how the human microbiome relates to health and disease.
Figures
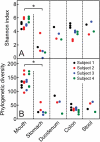
Figure 1. Alpha diversity of each sample.
(a) Shannon diversity and (b) phylogenetic diversity (PD) at each body site for all four subjects, where points represent a sample. Five iterations of rarefied subsets of 200,000 sequences from each sample were used to calculate the values for both metrics and the average was plotted. Asterisks indicate statistical significance (Kruskal-Wallis test, p < 0.05).
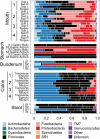
Figure 2. Genera represented in the digestive tracts of four subjects (S1–S4).
Bars delineate unique genera and are coloured with the phylum level assignment for each group. A complete list of sequence counts for each phylum and generus grouped by sample are presented in Supplemental tables 3 and 4, respectively. Low abundance phyla comprising the “Other” category include Acidobacteria, Chloroflexi, Deinococcus-Thermus, Euryarchaeota, Lentisphaerae, Planctomycetes, Synergistetes and Tenericutes.
Figure 3. Contribution of different taxonomic groups to separation of samples based on phylogenetic information.
The contribution of each group is represented by the size of the circles (grey) overlayed onto a PCoA of unweighted UniFrac distances for all samples within the oral and digestive tract. Panels (a–d) represent variations in sample colouration to highlight potential relationships between sample clustering and metadata.
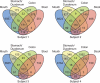
Figure 4. Venn diagrams demonstrating 97% OTU cluster overlap within broad sampling regions.
Numbers correspond to unique OTU clusters within a subset. To highlight shared OTUs, singleton clusters were removed before analysis.
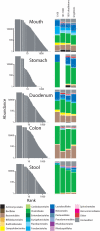
Figure 5. Rank abundance plots and proportional taxonomic bar plots for sequences from Subject 1 pooled within various body sites and assigned to order.
To demonstrate taxonomic distribution at decreasing ranks, the data were split according to different logarithmic abundance ranges as appropriate. The low abundance orders comprising the “Other” category included Synergistales, Halanaerobiales, Mycoplasmatales, Xanthomonadales, Sphingobacteriales, Caulobacterales, Desulfobacterales, Legionellales, Oceanospirillales, Deinococcales, Methanobacteriales, Myxococcales, Anaerolineales, Methylophilales, Chromatiales, Thermales, Bdellovibrionales, Desulfuromonadales, Solirubrobacterales, Methanomicrobiales, Planctomycetales, Methylococcales, Anaeroplasmatales, Coriobacteriales, Desulfovibrionales, Rhizobiales, Rhodocyclales, Sphingomonadales, Victivallales. Plots for subjects 2–4 are available in Figures S5.
Figure 6. Abundance of V3 regions of 16S rRNA genes of targeted organisms within all samples.
The entire sequence dataset was queried for type strains of Streptococcus mutans, Treponema denticola, Clostridium difficile and Faecalibacterium prausnitzii. For results from additional target organisms, see Table S6.
Similar articles
- Bacterial diversity in the rumen of Indian Surti buffalo (Bubalus bubalis), assessed by 16S rDNA analysis.
Pandya PR, Singh KM, Parnerkar S, Tripathi AK, Mehta HH, Rank DN, Kothari RK, Joshi CG. Pandya PR, et al. J Appl Genet. 2010;51(3):395-402. doi: 10.1007/BF03208869. J Appl Genet. 2010. PMID: 20720314 - Tissue-Associated Bacterial Alterations in Rectal Carcinoma Patients Revealed by 16S rRNA Community Profiling.
Thomas AM, Jesus EC, Lopes A, Aguiar S Jr, Begnami MD, Rocha RM, Carpinetti PA, Camargo AA, Hoffmann C, Freitas HC, Silva IT, Nunes DN, Setubal JC, Dias-Neto E. Thomas AM, et al. Front Cell Infect Microbiol. 2016 Dec 9;6:179. doi: 10.3389/fcimb.2016.00179. eCollection 2016. Front Cell Infect Microbiol. 2016. PMID: 28018861 Free PMC article. - Assessment of microbial diversity along the feline intestinal tract using 16S rRNA gene analysis.
Ritchie LE, Steiner JM, Suchodolski JS. Ritchie LE, et al. FEMS Microbiol Ecol. 2008 Dec;66(3):590-8. doi: 10.1111/j.1574-6941.2008.00609.x. FEMS Microbiol Ecol. 2008. PMID: 19049654 - Defining the human microbiome.
Ursell LK, Metcalf JL, Parfrey LW, Knight R. Ursell LK, et al. Nutr Rev. 2012 Aug;70 Suppl 1(Suppl 1):S38-44. doi: 10.1111/j.1753-4887.2012.00493.x. Nutr Rev. 2012. PMID: 22861806 Free PMC article. Review. - Worlds within worlds: evolution of the vertebrate gut microbiota.
Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Ley RE, et al. Nat Rev Microbiol. 2008 Oct;6(10):776-88. doi: 10.1038/nrmicro1978. Nat Rev Microbiol. 2008. PMID: 18794915 Free PMC article. Review.
Cited by
- Mapping the New World of Necrotizing Enterocolitis (NEC): Review and Opinion.
Gordon P, Christensen R, Weitkamp JH, Maheshwari A. Gordon P, et al. EJ Neonatol Res. 2012;2(4):145-172. EJ Neonatol Res. 2012. PMID: 23730536 Free PMC article. - The Effects of Helicobacter pylori Infection on Microbiota Associated With Gastric Mucosa and Immune Factors in Children.
Zheng W, Miao J, Luo L, Long G, Chen B, Shu X, Gu W, Peng K, Li F, Zhao H, Botchway BOA, Fang M, Jiang M. Zheng W, et al. Front Immunol. 2021 Mar 24;12:625586. doi: 10.3389/fimmu.2021.625586. eCollection 2021. Front Immunol. 2021. PMID: 33841407 Free PMC article. - A toolbox for the comprehensive analysis of small volume human intestinal samples that can be used with gastrointestinal sampling capsules.
Rios-Morales M, van Trijp MPH, Rösch C, An R, Boer T, Gerding A, de Ruiter N, Koehorst M, Heiner-Fokkema MR, Schols HA, Reijngoud DJ, Hooiveld GJEJ, Bakker BM. Rios-Morales M, et al. Sci Rep. 2021 Apr 14;11(1):8133. doi: 10.1038/s41598-021-86980-y. Sci Rep. 2021. PMID: 33854074 Free PMC article. - A literature review on the potential clinical implications of streptococci in gastric cancer.
Zi M, Zhang Y, Hu C, Zhang S, Chen J, Yuan L, Cheng X. Zi M, et al. Front Microbiol. 2022 Oct 26;13:1010465. doi: 10.3389/fmicb.2022.1010465. eCollection 2022. Front Microbiol. 2022. PMID: 36386672 Free PMC article. Review. - Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment.
Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, Bousvaros A, Korzenik J, Sands BE, Xavier RJ, Huttenhower C. Morgan XC, et al. Genome Biol. 2012 Apr 16;13(9):R79. doi: 10.1186/gb-2012-13-9-r79. Genome Biol. 2012. PMID: 23013615 Free PMC article.
References
- Mazmanian S. K., Liu C. H., Tzianabos A. O. & Kasper D. L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118 (2005). - PubMed
- Bäckhed F., Ley R. E., Sonnenburg J. L., Peterson D. A. & Gordon J. I. Host-bacterial mutualism in the human intestine. Science 307, 1915–1920 (2005). - PubMed
- Beck J. D. et al.. Periodontal disease and coronary heart disease: a reappraisal of the exposure. Circulation 112, 19–24 (2005). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources