RAE-1, a novel PHR binding protein, is required for axon termination and synapse formation in Caenorhabditis elegans - PubMed (original) (raw)
RAE-1, a novel PHR binding protein, is required for axon termination and synapse formation in Caenorhabditis elegans
Brock Grill et al. J Neurosci. 2012.
Abstract
Previous studies in Caenorhabditis elegans showed that RPM-1 (Regulator of Presynaptic Morphology-1) regulates axon termination and synapse formation. To understand the mechanism of how rpm-1 functions, we have used mass spectrometry to identify RPM-1 binding proteins, and have identified RAE-1 (RNA Export protein-1) as an evolutionarily conserved binding partner. We define a RAE-1 binding region in RPM-1, and show that this binding interaction is conserved and also occurs between Rae1 and the human ortholog of RPM-1 called Pam (protein associated with Myc). rae-1 loss of function causes similar axon and synapse defects, and synergizes genetically with two other RPM-1 binding proteins, GLO-4 and FSN-1. Further, we show that RAE-1 colocalizes with RPM-1 in neurons, and that rae-1 functions downstream of rpm-1. These studies establish a novel postmitotic function for rae-1 in neuronal development.
Figures
Figure 1.
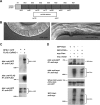
PHR proteins bind to RAE-1. A, A schematic of the CeRAE-1 protein which is composed of an N-terminal domain and 7 WD repeats. Shown below is the segment of the RAE-1 protein that is deleted by rae-1(tm2784). B, Shown is a 40× DIC image of the vulval region of a wild-type animal and a rae-1(tm2784) mutant. Note the absence of embryos and oocytes, and the protruding vulva in the rae-1 mutant, which can be used to identify these animals. C, coIP of RPM-1::GFP with FLAG::RAE-1. RPM-1::GFP was expressed transgenically alone or in combination with FLAG::RAE-1 in the neurons of C. elegans. D, coIP of myc-Pam and GFP-Rae1. Myc-tagged human Pam and GFP-tagged rat Rae1 or GFP-rat PP2Cα (a protein of similar size to Rae1) were coexpressed in 293 cells. For B and C, a representative of at least three independent experiments is shown. For B, similar results were obtained using a minimum of two transgenic lines.
Figure 2.
Identification of a Rae1 binding motif in the PHR proteins. A, Schematic representation of human Pam and C. elegans RPM-1. RLD (RCC-1 like domain), PHR (Pam/Highwire/RPM-1 family-specific domains), RBD (Rae1 binding domain), MBD (myc binding domain), NLS (nuclear localization signal), and RING (RING-H2 ubiquitin ligase domain). Shown below is a sequence alignment of the known Rae1 binding protein, Nup98, and the PHR proteins. Residues highlighted in blue are conserved between Nup98 and other previously described Rae1 binding proteins. Residues in Nup98 that are underlined are essential for binding to Rae1. B, coIP of the Pam RBD or the RPM-1 RBD fused to GFP with Flag-tagged rat Rae1. C, Identification of point mutations in the RPM-1 RBD that reduce binding to FLAG-Rae1. D, coIP of RPM-1::GFP or RPM-1::GFP (VIR to AAA) point mutant with FLAG::RAE-1 from transgenic C. elegans protein lysates. Note that the RPM-1::GFP (VIR to AAA) point mutant has greatly reduced binding to FLAG::RAE-1 compared with wild-type RPM-1::GFP. A representative of at least three independent experiments is shown for B–D. For D, at least two independently derived transgenic lines were analyzed for each genotype.
Figure 3.
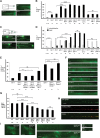
_rae-1_−/− mutants have defects in axon termination and synapse formation. A, C, Mechanosensory neurons (ALM and PLM) were visualized using muIs32. A, ALM axon termination defects in _rpm-1_−/− and _rae-1_−/− animals. B, Quantitation of ALM axon termination defects for the indicated genotypes. C, PLM axon termination defects in _rpm-1_−/− and _rae-1_−/− animals. The arrowhead highlights an example of the hook and overextension (hook) defect in rpm-1 mutants and the arrow highlights the less penetrant overextension defect in _rpm-1_−/− mutants. D, Quantitation of PLM axon termination defects for the indicated genotypes. E, Expression of C. elegans rae-1 or a cDNA encoding rat Rae1 in the mechanosensory neurons rescues the axon termination defects (hook) in the PLM neurons of rae-1_−/−;fsn-1_−/− animals. Overexpression of rae-1 was also sufficient to partially rescue defects in _rpm-1_−/− animals. For all extrachromosomal arrays, data from 4 to 10 independently derived transgenic lines was pooled. F, The presynaptic terminals of the DD neurons (dorsal cord) were visualized using SNB-1::GFP. Arrows highlight examples of gaps where presynaptic terminals are absent. G, Quantitation of synapse formation defects in the DD neurons. H, Confocal microscopy was used to visualize the dorsal cord of transgenic animals that express RPM-1::GFP (juIs77) and mCherry::RAE-1 (bggEx76) in the GABAergic motor neurons using the unc-25 promoter. mCherry::RAE-1 forms discrete puncta in the dorsal cord that colocalize with RPM-1::GFP puncta in the perisynatic zone of presynaptic terminals. I, Epifluorescent microscopy was used to visualize RPM-1::GFP driven by its own promoter (juIs58) in the nerve ring, dorsal cord, and SAB neurons of rae-1+/− or _rae-1_−/− animals. No difference in the levels and localization of RPM-1::GFP were observed in rae-1 mutants compared with heterozygous animals. Analysis of axon termination was done at 23°C, and analysis of synapse formation was done at 25°C. Scale bars are 10 μm. For B and D, n is the number of individual mechanosensory neurons that were scored. For G, n is the number of images of the dorsal cord that were quantified. For axon termination defects, error bars represent the SE, and significance was determined using a two-tailed Fisher's exact test. For synapse formation defects, error bars represent SEM, and significance was determined using a t test. **p < 0.005, ***p < 0.001, and ns=not significant.
Figure 4.
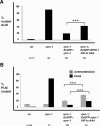
Binding of RAE-1 to RPM-1 is necessary for RPM-1 to be fully functional. Transgenic animals were generated on an _rpm-1_−/− background to quantify rescue of rpm-1 (lf) defects. Rescue was analyzed for transgenic expression of wt RPM-1::GFP (Exrpm-1::GFP), or a point mutant of RPM-1::GFP (Exrpm-1::GFP VIR to AAA). A, ALM axon termination defects were quantified for the indicated genotypes. B, PLM axon termination defects were quantified for the indicated genotypes. Note, that data from 4–6 independently derived transgenic lines was pooled for this analysis, and n is the number of ALM or PLM neurons that were scored. A two-tailed Fisher exact test was used to determine significance. ***p < 0.0005.
Similar articles
- Identification of a peptide inhibitor of the RPM-1 · FSN-1 ubiquitin ligase complex.
Sharma J, Baker ST, Turgeon SM, Gurney AM, Opperman KJ, Grill B. Sharma J, et al. J Biol Chem. 2014 Dec 12;289(50):34654-66. doi: 10.1074/jbc.M114.614065. Epub 2014 Oct 17. J Biol Chem. 2014. PMID: 25326385 Free PMC article. - Defining Minimal Binding Regions in Regulator of Presynaptic Morphology 1 (RPM-1) Using Caenorhabditis elegans Neurons Reveals Differential Signaling Complexes.
Baker ST, Grill B. Baker ST, et al. J Biol Chem. 2017 Feb 10;292(6):2519-2530. doi: 10.1074/jbc.M116.748004. Epub 2016 Dec 15. J Biol Chem. 2017. PMID: 27979965 Free PMC article. - C. elegans RPM-1 regulates axon termination and synaptogenesis through the Rab GEF GLO-4 and the Rab GTPase GLO-1.
Grill B, Bienvenut WV, Brown HM, Ackley BD, Quadroni M, Jin Y. Grill B, et al. Neuron. 2007 Aug 16;55(4):587-601. doi: 10.1016/j.neuron.2007.07.009. Neuron. 2007. PMID: 17698012 - The PHR proteins: intracellular signaling hubs in neuronal development and axon degeneration.
Grill B, Murphey RK, Borgen MA. Grill B, et al. Neural Dev. 2016 Mar 23;11:8. doi: 10.1186/s13064-016-0063-0. Neural Dev. 2016. PMID: 27008623 Free PMC article. Review. - Unraveling the mechanisms of synapse formation and axon regeneration: the awesome power of C. elegans genetics.
Jin Y. Jin Y. Sci China Life Sci. 2015 Nov;58(11):1084-8. doi: 10.1007/s11427-015-4962-9. Epub 2015 Nov 13. Sci China Life Sci. 2015. PMID: 26563175 Free PMC article. Review.
Cited by
- Advances in the understanding of nuclear pore complexes in human diseases.
Li Y, Zhu J, Zhai F, Kong L, Li H, Jin X. Li Y, et al. J Cancer Res Clin Oncol. 2024 Jul 30;150(7):374. doi: 10.1007/s00432-024-05881-5. J Cancer Res Clin Oncol. 2024. PMID: 39080077 Free PMC article. Review. - Ubiquitin ligase and signalling hub MYCBP2 is required for efficient EPHB2 tyrosine kinase receptor function.
Chang C, Banerjee SL, Park SS, Zhang XL, Cotnoir-White D, Opperman KJ, Desbois M, Grill B, Kania A. Chang C, et al. Elife. 2024 Jan 30;12:RP89176. doi: 10.7554/eLife.89176. Elife. 2024. PMID: 38289221 Free PMC article. - Non-FG mediated transport of the large pre-ribosomal subunit through the nuclear pore complex by the mRNA export factor Gle2.
Occhipinti L, Chang Y, Altvater M, Menet AM, Kemmler S, Panse VG. Occhipinti L, et al. Nucleic Acids Res. 2013 Sep;41(17):8266-79. doi: 10.1093/nar/gkt675. Epub 2013 Jul 31. Nucleic Acids Res. 2013. PMID: 23907389 Free PMC article. - Insights into dynamic mitotic chromatin organization through the NIMA kinase suppressor SonC, a chromatin-associated protein involved in the DNA damage response.
Larson JR, Facemyer EM, Shen KF, Ukil L, Osmani SA. Larson JR, et al. Genetics. 2014 Jan;196(1):177-95. doi: 10.1534/genetics.113.156745. Epub 2013 Nov 8. Genetics. 2014. PMID: 24214344 Free PMC article. - Optimized protocol for in vivo affinity purification proteomics and biochemistry using C. elegans.
Desbois M, Pak JS, Opperman KJ, Giles AC, Grill B. Desbois M, et al. STAR Protoc. 2023 Jun 7;4(2):102262. doi: 10.1016/j.xpro.2023.102262. Online ahead of print. STAR Protoc. 2023. PMID: 37294631 Free PMC article.
References
- Blevins MB, Smith AM, Phillips EM, Powers MA. Complex formation among the RNA export proteins Nup98, Rae1/Gle2, and TAP. J Biol Chem. 2003;278:20979–20988. - PubMed
- Blower MD, Nachury M, Heald R, Weis K. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 2005;121:223–234. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 NS035546/NS/NINDS NIH HHS/United States
- R01 NS072129-02/NS/NINDS NIH HHS/United States
- R01 NS072129/NS/NINDS NIH HHS/United States
- R01 NS035546/NS/NINDS NIH HHS/United States
- R01 NS039471/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous